- Title
-
Cooperation of Mtmr8 with PI3K regulates actin filament modeling and muscle development in zebrafish
- Authors
- Mei, J., Li, Z., and Gui, J.F.
- Source
- Full text @ PLoS One
|
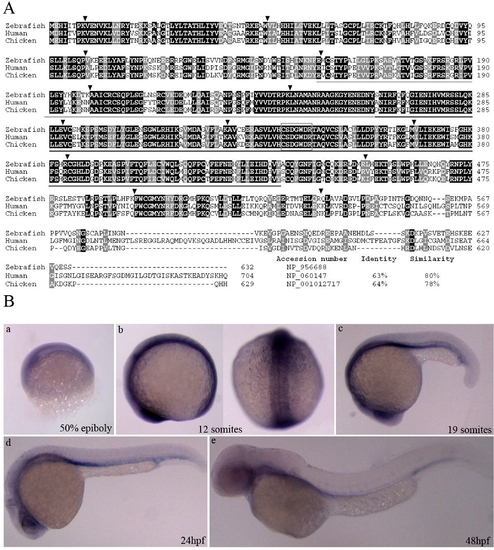
Sequence comparison and expression pattern of the deduced zebrafish Mtmr8. (A) Amino acid alignment of Mtmr8 between zebrafish with human and chicken. Similar and identical amino acids are highlighted in grey and black boxes. And the percentages of identities and similarities in Mtmr8 were shown compared zebrafish with others. Arrowheads indicate the location of introns and are flanked by the corresponding exon numbers. The region encompassing the Myotub-related and PTPc_DSPc domain is underlined in grey and black lines. The rectangular box indicates the CX5R active site motif of enzymatically active members in the MTM family. (B) Expression pattern of zebrafish Mtmr8. Whole-mount RNA in situ hybridization were performed using a Mtmr8 specific antisense riboprobe on embryos at the indicated stages. The arrows indicate the signals in the anterior and head. Embryos in panels are lateral view with the animal pole toward the top, and the right picture of panel b is dorsal view. The embryos in other panels are lateral views, with dorsal toward the top and anterior to the left. All scale bars are 100 μm. EXPRESSION / LABELING:
|
|
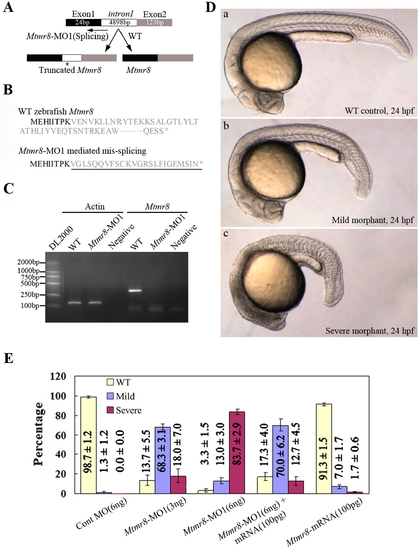
Targeted knockdown of Mtmr8 using splicing morpholino in zebrafish embryos. (A) Diagram of splicing junction morpholino targeted against Mtmr8 exon-intron boundary. (B) Amino acid sequences of wild type Mtmr8 and Mtmr8-MO1 mediated mis-splicing Mtmr8. Sequencing of the RT-PCR products revealed the mis-splicing transcript leading to a premature stop (asterisk) and causing a truncation in the protein (exon 1 in bold, the other exons in plain, and intron in plain and underlined). (C) RT-PCR detection of Mtmr8 transcript at 24hpf in WT and Mtmr8-MO1 (6 ng) morpholino-injected embryos, comparing cryptic spliced transcript in the morpholino injected embryos to the cDNA PCR products. (D) Live morphology of WT control zebrafish embryo (a) and Mtmr8-MO1 knockdown embryo (b, c) at 24hpf. Injection volume was about 2 nL at 1-cell stage embryos. All scale bars are 100 μm. (E) Statistical data of three independent experiments on Mtmr8 knockdown as well as its overexpression and Mtmr8 mRNA rescue. Results are represented as mean±SD of three separate experiments (60 embryos in each experiment). |
|
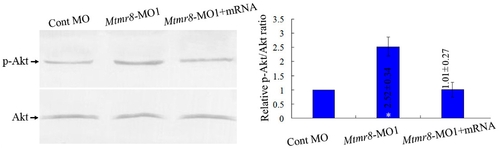
Phosphatase activity of zebrafish Mtmr8. Different extracts from Cont MO embryos, Mtmr8 morphants and Mtmr8 morphants co-injected with Mtmr8 mRNA were prepared at 24 hpf (30 embryos/group) and subjected to Western blot detection with anti-Akt antibody or anti-phospho-Akt (pAkt) antibody. The corresponding histograms (right panels) plot the relative Ser(P)473-Akt to total Akt ratio (Relative p-Akt/Akt ratio), determined by band intensities that were analyzed by Scion software, with the ratio in experiment extracts normalized to the ratio determined in Cont MO injected embryo extracts. Results represent mean±SD of three separate experiments. *P<0.05. |
|
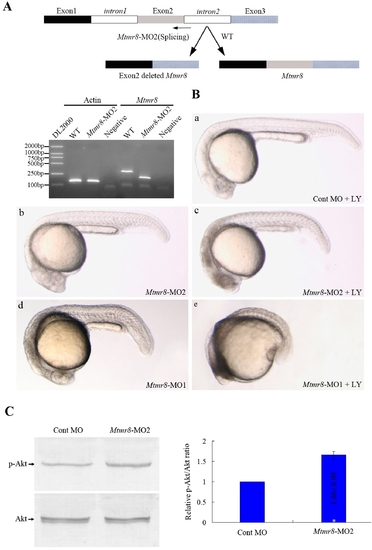
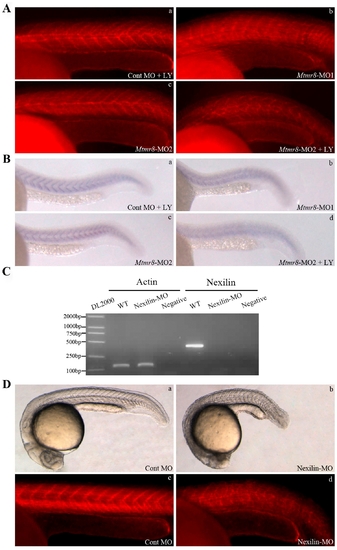
PH/G domain of Mtmr8 is required to cooperate with PI3K regulating embryo development and essential for the phosphatase activity of zebrafish Mtmr8 gene. (A) Splice junction morpholino targeted against Mtmr8 exon-intron boundary resulted to loss of PH/G domain. RT-PCR of Mtmr8 transcript at 24hpf in WT and Mtmr8-MO2 (6 ng) morpholino-injected embryos, comparing cryptic spliced transcript in the morpholino injected embryos to the cDNA PCR products. (B) Embryo phenotypes observed under the microscope. LY represents the PI3K inhibitor, LY294002. Each picture represents typical results out of three separate experiments (30 embryos in each experiment). All scale bars are 100 μm. (C) Immunoblot of different extracts from Cont MO embryos and Mtmr8-MO2 morphants at 24 hpf (30 embryos/group). Relative p-Akt/Akt ratio was determined by band intensities that were analyzed by Scion software, with the ratio in experiment extracts normalized to the ratio determined in Cont MO injected embryo extracts. Results represent mean±SD of three separate experiments. *P<0.05. |
|
Mtmr8 cooperates with PI3K regulating actin modeling. (A) Staining of embryos with phalloidin (F-actin) at 24hpf. (B) Nexilin mRNA expression at 24 hpf. (C) RT-PCR of Nexilin transcript at 24hpf in WT and Nexilin-MO (3 ng) morpholino-injected embryos, comparing cryptic spliced transcript in the morpholino injected embryos to the cDNA PCR products. (D) The effects of Nexilin knockdown on embryo development (a, b) and F-actin (c, d). The pictures are representative of at least three experiments (30 embryos in each experiment). All scale bars are 100 μm. EXPRESSION / LABELING:
|
|
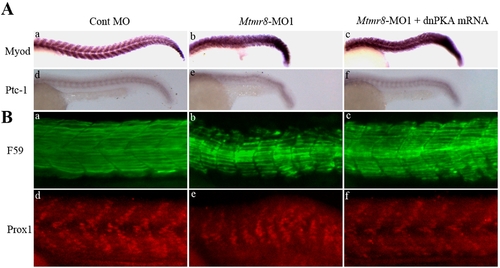
The effect of Mtmr8 knockdown on hedgehog pathway and slow muscle development. (A) The expression of myod and ptc1 at 24hpf in the Cont MO (a, d), Mtmr8-MO1 morphant (b, e) and it injected with dnPKA mRNA (c, f). (B) Whole-mount staining of 24 hpf embryo with F59 antibody (green) and Prox1 antibody (red) and to reveal slow muscle cells. The pictures are representative of at least three experiments (30 embryos in each experiment). All scale bars are 100 μm. PHENOTYPE:
|
|
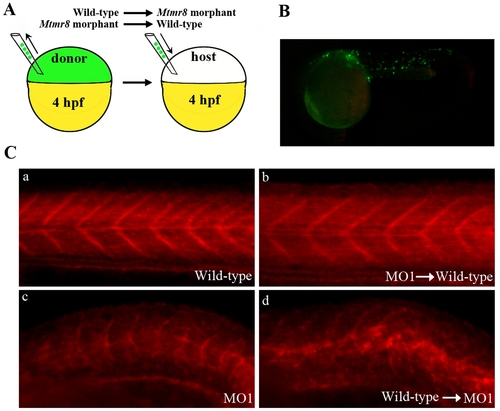
Mtmr8 controls actin modeling non-cell-autonomously. (A) Schematic depiction of cell transplant experiments. (B) Representative picture of a chimeric embryo at 24 hpf. The labeling cells were mostly located in somites of trunk and tail regions. (C) The effects of cell transplantation on F-actin. |