- Title
-
Crystal Structure of a Full-Length beta-Catenin
- Authors
- Xing, Y., Takemaru, K., Liu, J., Berndt, J.D., Zheng, J.J., Moon, R.T., and Xu, W.
- Source
- Full text @ Structure
|
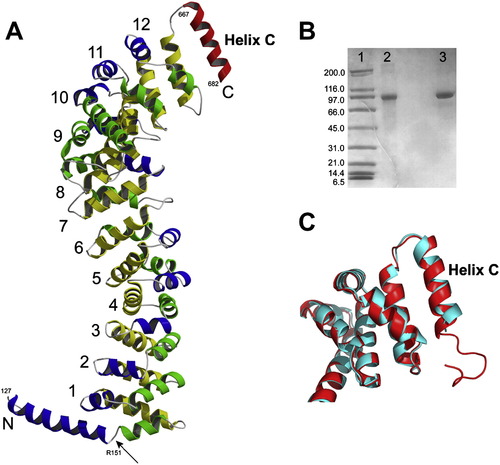
Full-Length Zebrafish β-Catenin Crystal Structure (A) Crystal structure of full-length zebrafish β-catenin. The color scheme of β-catenin armadillo repeats is the same as that in Figure 1. The C-terminal helix (helix C) is colored in red. The N-terminal helix is colored in blue and green with an arrow pointing to the kink. All β-catenin residue numbers shown in this figure are these of corresponding residue numbers of human β-catenin. (B) SDS-gel analysis of the dissolved full-length zebrafish β-catenin crystals. Lane 1, molecular weight marker. Lane 2, dissolved crystals. Lane 3, purified full-length zebrafish β-catenin. (C) Structural superposition between human β-catenin-R1C (red) and full-length zebrafish β-catenin (cyan) around helix C. |
|
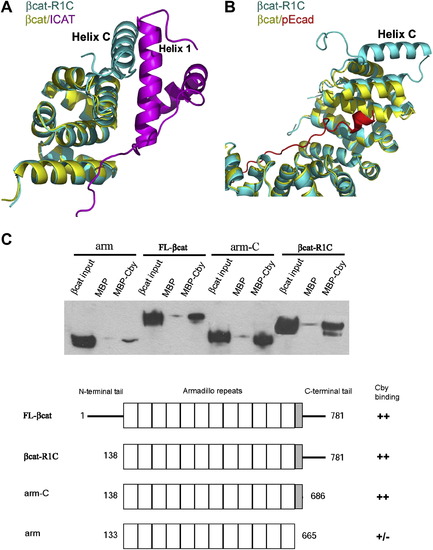
The Role of Helix C in β-Catenin Protein-Protein Interactions (A) Potential interactions between β-catenin helix C and ICAT. β-catenin-R1C (βcat-R1C, cyan) is superimposed onto β-catenin/ICAT (βcat/ICAT, yellow and magenta), based on the Cα's of β-catenin armadillo repeats 10–12 (residues 563–663). (B) Helix C is unlikely to affect the interaction between the phophorylated E-cadherin and β-catenin armadillo repeats. β-catenin-R1C (βcat-R1C, cyane) is superimposed with β-catenin/phospho-Ecadherin (βcat/pEcad, yellow and red, PDB code: 1I7W) based on all twelve armadillo repeats. (C) The helix C is required for β-catenin to interact with Chibby. Purified MBP-Chibby protein was immobilized to amylose beads; purified β-catenin and β-catenin fragments were tested for their binding to immobilized MBP-Chibby. The total input and bound β-catenin and β-catenin fragments were visualized by Western blot. |
|
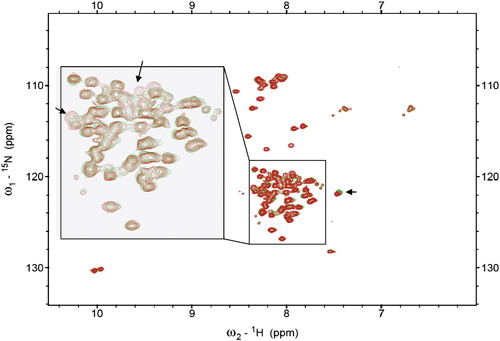
NMR Analysis of Potential Intramolecular Interactions of β-Catenin NMR studies suggest highly dynamic interactions between the C-terminal tail and the β-catenin N-R12 fragment 15N-TROSY spectra of the 15N-labeled β-catenin(687–781) alone (green) and with unlabeled β-catenine(1–686) (red). Poor dispersion of the resonance indicates that β-catenin(687–781) is unstructured in the absence or presence of β-catenine(1–686). Some minor chemical shift perturbations, as indicated by the arrows, suggest that β-catenin(687–781) may interact with β-catenine(1–686) in a highly dynamic manner. |
|
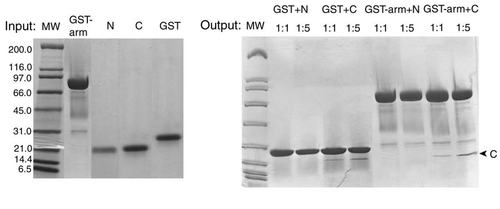
In Vitro GST Pull-Down Assays between Terminal Domains and Armadillo Repeat Domain of β-Catenin GST-tagged armadillo repeat domain (GST-arm) does not pull down significant amount of β-catenin N-terminal domain (N) or C-terminal domain (C) in comparison with the blank experiment done with GST. Protein bands in the gel are visualized by Coomassie Brilliant Blue. |
Reprinted from Structure (London, England : 1993), 16(3), Xing, Y., Takemaru, K., Liu, J., Berndt, J.D., Zheng, J.J., Moon, R.T., and Xu, W., Crystal Structure of a Full-Length beta-Catenin, 478-487, Copyright (2008) with permission from Elsevier. Full text @ Structure