- Title
-
tortuga refines Notch pathway gene expression in the zebrafish presomitic mesoderm at the post-transcriptional level
- Authors
- Dill, K.K., and Amacher, S.L.
- Source
- Full text @ Dev. Biol.
|
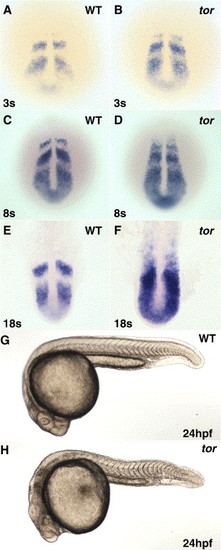
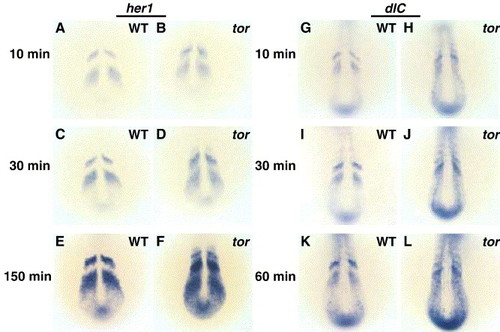
Oscillating her1 mRNA expression is disrupted in tortuga mutants, but somite boundary formation occurs normally. In situ hybridization in wild type (A, C, E) and tor mutant (B, D, F) embryos indicates that her1 is misexpressed in tor mutant embryos from the beginning of segmentation, and defects get progressively worse as segmentation progresses. Panels are dorsal views, anterior top, with the somite stages indicated in lower left and phenotype indicated in the upper right of each panel. (G, H) Morphological examination shows that tor mutant embryos have segmental pattern, although posterior somites appear more U-shaped in tor mutant embryos compared to the wild type chevron shape. Other defects apparent in this view are head necrosis and reduced posterior yolk. Panels are lateral views of embryos at 24 hpf. EXPRESSION / LABELING:
|
|
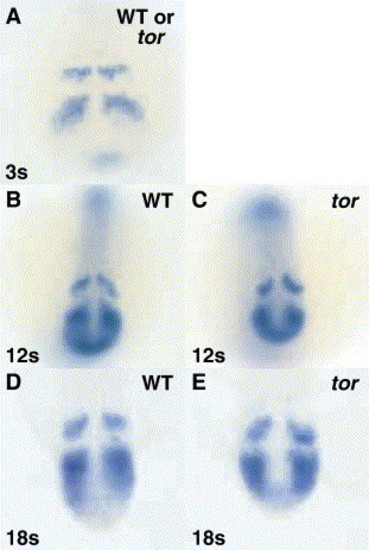
tortuga regulates her1 at the post-transcriptional level. Embryos were hybridized with an in situ hybridization probe that binds the third intron of her1 and thus detects nascent and/or unprocessed transcript. At the 3 somite stage, mutant embryos in a clutch derived from tor+/- parents cannot be distinguished from wild type siblings (A). At 12 and 18 somite stages, tor mutant embryos were sorted from their wild type siblings by morphological criteria before hybridization; again, no differences in the pattern of her1 transcription in wild type and tor mutant embryos are observed (B–E). Panels are dorsal views, anterior top. EXPRESSION / LABELING:
|
|
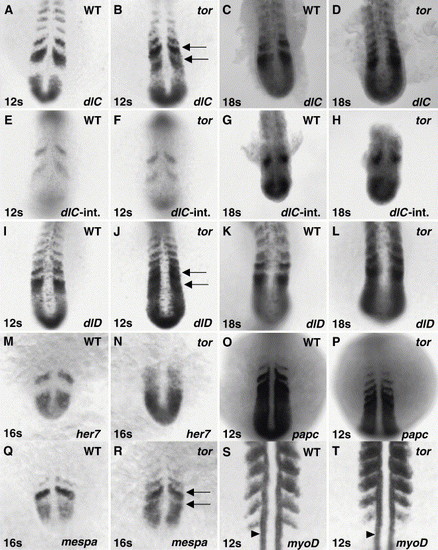
tor mutant embryos exhibit distinct disruptions in the expression of Notch pathway genes and subtle disruptions in somite polarity. (A–D and I–N) Dynamically expressed Notch pathway genes, deltaC (dlC), deltaD (dlD), and her7, are misexpressed in tor mutant embryos in a manner similar to her1, with normal stripes and ectopic expression between stripes, although the timing of disruption is delayed relative to her1. Arrows in Panels B and J indicate ectopic expression in tor mutant PSM. (E–H) An intronic probe for deltaC was used to detect nascent and/or unprocessed transcript. At 12 and 18 somite stages, the pattern of deltaC transcription in tor mutant embryos appears normal (compare to exonic probe hybridization in Panels A–D). (O–T) Markers of somite polarity exhibit subtle disruptions in tor mutant embryos. In mutant embryos, papc expression is normal within the PSM, but expression is detected longer after somites form; after 3 h of coloration detection, we see 3 to 4 stripes of papc expression in wild type embryos (O) and 4 to 6 stripes in tor mutant embryos (P). The polarized expression of mespa in the presumptive anterior compartments is apparent in wild type and tor mutant embryos (Q, R); however, low levels of transcript are also detected in the posterior compartment of presumptive somites in mutant embryos (arrows in R). The posterior polarity of myoD is maintained in tor mutant embryos, although myoD stripes are not as distinct as in wild type, and some transcript is detected in the anterior compartment (S,T). myoD expression in slow muscle precursors (arrowheads in S and T) is normal in tor mutant embryos. Panels are dorsal views, anterior top. Genotype, stage, and probe are indicated on each panel. EXPRESSION / LABELING:
|
|
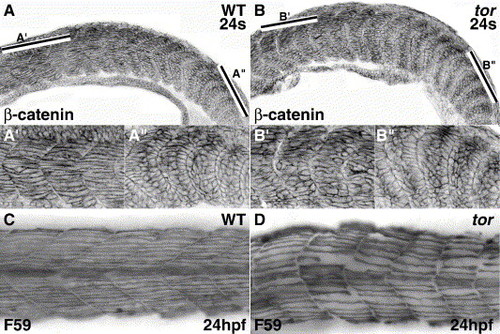
Somite boundaries form normally, but defects appear later, as tor mutant somitic cells mature into skeletal muscle. Confocal micrographs of β-catenin expression show similar cubiodal morphology of cells at newly formed/forming boundaries in wild type (A, A″) and tor mutant (B, B″) embryos. In wild type embryos, myofibers in anterior somites have extended across the entire myotome (A′), while myofibers in anterior mutant somites have not completed elongation (B′). The bars in Panels A and B indicate the respective regions magnified in the prime and double prime panels. Slow muscle myosin expression (C, D) shows the abnormal morphology of slow muscle in tor mutant embryos; slow fibers in mutant embryos are irregularly spaced, do not extend in a straight trajectory, and boundaries between the myotomes are irregular. Panels are lateral views, anterior to the left. Genotype and stage are indicated on the panels. |
|
Ectopic expression of her1 and deltaC in tor mutant embryos is visible at times when wild type expression domains first become apparent. Embryos from the same clutch were fixed at 8 somites, hybridized with her1 (A–F) or deltaC (G–L) riboprobes, and processed in parallel coloration reactions. After 10 min, wild type expression domains are apparent (A, G). tor mutant embryos exhibit similar staining intensity in the wild type expression domains (stripes) with lower staining intensity in the ectopic interstripe domains (B, H). After longer incubations, both the wild type and ectopic expression domains show progressively darker staining intensity as the coloration reaction approaches saturation (B–F and H–L). Panels are dorsal views, anterior top. Genotype is indicated on each panel, and the duration of the coloration reaction is indicated to the left. Embryos were fixed before they could be separated by morphological criteria, and the ratio of presumptive wild type to tor mutant embryos was consistent with the expected 3:1 Mendelian ratio. |
|
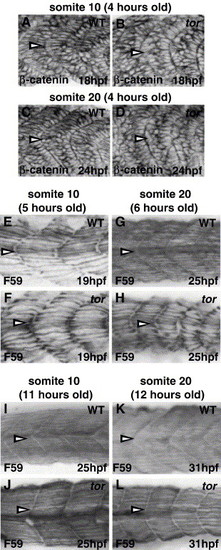
Muscle fiber defects (elongation and slow fiber morphology) in tor mutant embryos are similar in anterior and posterior somites. Embryos were stained with β-catenin to compare fast muscle cell elongation in anterior (somite 10; A, B) and posterior (somite 20; C, D) somites. In wild type embryos, many fast muscle cells have initiated elongation 4 h after somite formation, regardless of position along the anterior–posterior axis (A, C; medial sections are 4–5 μM lateral to the notochord). In contrast, most tor mutant fast muscle cells at the same developmental stage remain mesenchymal, and the elongation delay is similar in somite 10 (anterior) and somite 20 (posterior) (B, D). (E–L) Slow muscle myosin (F59) expression reveals that slow muscle cell development is similar in anterior and posterior somites of tor mutant embryos. Approximately 5–6 h after somite formation, wild type slow muscle fibers reveal the basic metameric pattern (E, G), gaps appear in tor mutant somites, and the organization and morphology of slow muscle fibers appear similar in somite 10 and somite 20 (compare Panel F to H). Slow muscle defects recover in tor mutant somite/myotomes that formed 11–12 h earlier, and it is difficult to distinguish between wild type and mutant myotome based on slow muscle cell morphology alone (I–L), although tor mutant somite shape is distinctly U-shaped instead of chevron-shaped. All panels are lateral views with anterior to the left. Antibody and stage are indicated on each panel; somite number (indicated by arrowheads) and the age of that somite are indicated above each column. |
Reprinted from Developmental Biology, 287(2), Dill, K.K., and Amacher, S.L., tortuga refines Notch pathway gene expression in the zebrafish presomitic mesoderm at the post-transcriptional level, 225-236, Copyright (2005) with permission from Elsevier. Full text @ Dev. Biol.