- Title
-
valentino, a zebrafish gene required for normal hindbrain segmentation
- Authors
- Moens, C.B., Yan, Y.L., Appel, B., Force, A.G., and Kimmel, C.B.
- Source
- Full text @ Development
|
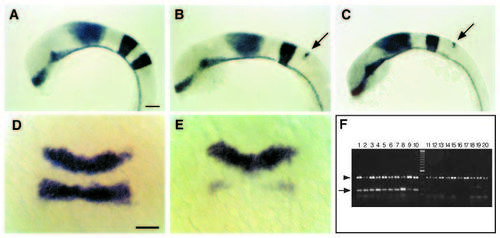
krox20 expression is disrupted in val- embryos early during hindbrain development. We screened 472 ENUmutagenized and 741 g- ray-mutagenized haploid genomes, and identified three alleles of valentino, one ENU-induced (valb337) and two g-rayinduced (valb361 and valb475). (A-C) Whole-mount RNA in situ hybridizations in lateral view showing expression of three genes, shh, en3 and krox20, in 18 h wild-type (A), valb337 (B) and valb361 (C) embryos. Anterior is to the left. In both alleles of valentino shown here, the r5 stripe of krox20 staining is reduced to a vestigial strip of expression in the dorsal hindbrain at the position of the r4-r5 boundary (arrow). (D,E) Dorsal views of whole-mount RNA in situs showing expression of krox20 in wild-type (D) and valb337 (E) embryos at the 2- to 3-somite stage (10J- 11 h). Anterior is to the top. In val- embryos, krox20 expression is already disrupted in the presumptive r5. (F) Following RNA in situ hybridization at the 2- to 3-somite stage, embryos from a cross between valb361/val+ individuals were sorted based on krox20 expression and then their genotype was determined by PCR using snail2, which is deleted in valb361 (see Materials and Methods). 10/10 individuals scored as wild-type were in fact wild type as determined by PCR (lanes 1-10) and 10/10 individuals scored as mutant were in fact mutant (lanes 11-20). Arrow: snail-2; arrowhead: nk2.2, an unlinked gene that is amplified from both wild-type and mutant DNA. Scale bars, 50 μm. EXPRESSION / LABELING:
|
|
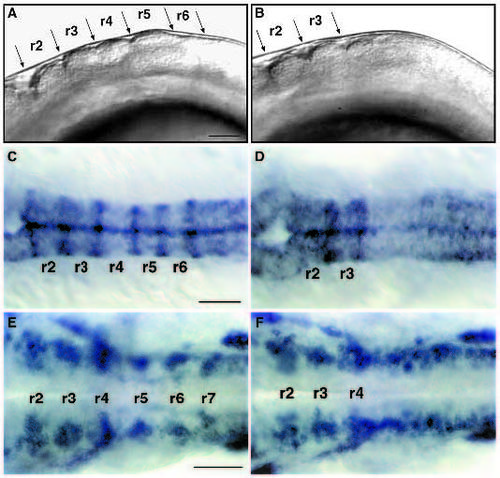
val- embryos lack segment boundaries and segmental patterns of neuronal differentiation posterior to rhombomere 4. (A,B) Lateral view of live 18 h wild-type (A) and val- (B) embryos. Anterior is to the left. In val- embryos, there are no visible rhombomere boundaries posterior to the r3-r4 boundary. (C,D) Dorsal views of RNA in situ hybridizations of wild-type (C) and val- (D) embryos at 18 h showing expression of mariposa in the rhombomere boundaries. No expression is observed posterior to the r3-r4 boundary in val- embryos. (E,F) Dorsal views of RNA in situ hybridizations of wildtype (E) and val- (F) embryos at 24 h showing expression of gap43 in clusters of early differentiating neurons laterally in each rhombomere. In val- embryos, this segmental pattern of gap43 staining is lost posterior to r4. This disrupted pattern of neuronal differentiation is also observed in val- embryos stained with the HNK-1 antibody (data not shown; Metcalfe et al., 1990; Trevarrow et al., 1990). Scale bars, 50 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
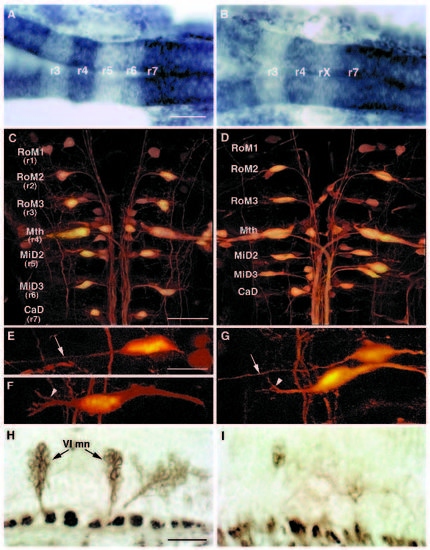
The hindbrain of val- embryos is reduced by the length of one rhombomere. (A,B) Dorsal view of 22 h wild-type (A) and val- (B) embryos showing expression of two genes: g13.1 in r4 and anterior to the r2-r3 boundary and hdc posterior to the r6-r7 boundary. Anterior is to the left. The distance between the posterior boundary of g13.1 expression in r4 and the anterior boundary of hdc expression is reduced by the length of approximately one rhombomere in the val- compared to the wild-type embryo. ‘rX’ refers to the region of that remains between r4 and r7. (C-G) Confocal images of 5-day old wild-type (C,E,F) and val- (D,G) embryos in which the hindbrain reticulospinal neurons are visualized by retrograde filling with lysinated rhodamine-dextran. Anterior is to the top. The names of individually identifiable neurons are indicated. In wild-type embryos, the RoM3 neurons lie in r3, Mauthner (Mth) in r4, MiD2 in r5, MiD3 in r6 and CaD in r7. In val- embryos, the average distance from Mth to CaD is reduced by the length of approximately one rhombomere, and the MiD2 and MiD3 neurons lie close together in the region of one rhombomere’s length between r4 and r7. (EG) Higher power confocal images of MiD2 and MiD3 cells in wild-type (E,F) and mutant (G) embryos. Arrows indicate the long, unbranched lateral dendrite characteristic of the MiD2cm cell, while arrowheads indicate the shorter, branched lateral dendrite characteristic of the MiD3cm cell. Note that, although the MiD2cm and MiD3cm cells are immediately adjacent to one another in the mutant embryo shown in G, the MiD2cm cell is still anterior to the MiD3cm cell. (H,I) Sagittal sections of 56 h wild-type (H) and val- (I) embryos stained with the zn-5 antibody, which labels the putative motor nuclei of the abducens nerve (VI) in rhombomeres 5 and 6. Anterior is to the left. These motor nuclei are largely absent in the val- embryo. The putative abducens motor nuclei differentiate relatively late during hindbrain development, since they first stain with zn-5 24 hours later than do the earlier differentiating hindbrain commissural neurons (Trevarrow et al., 1990). Scale bars, (A,B) 50 μm; (C,D) 50 μm; (E-G) 20 μm; (H,I) 20 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
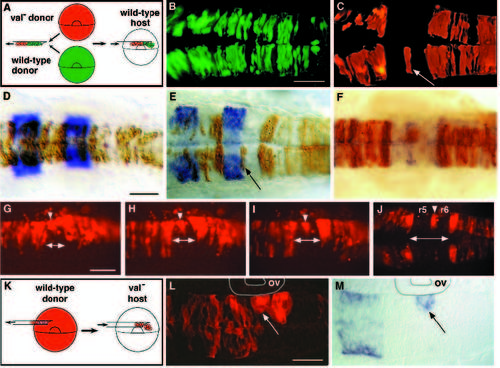
Mosaic analysis demonstrates that valentino is required for cells to contribute to either r5 or r6. (A) Schematic diagram of the experimental approach used for B-J. Rhodaminelabeled cells from a val- donor and fluoresceinlabeled cells from a wildtype donor were transplanted into the same unlabeled wild-type host at the early gastrula stage (see Materials and Methods). The distribution of labeled cells was then observed at 24 h of development. (B,C) Confocal images taken in dorsal view showing the distribution of wild-type (B) and val- cells (C) in the hindbrain region of the same wild-type host. val- cells are specifically excluded from a sharply defined region of the hindbrain, but are otherwise able to contribute to the entire brain and spinal cord. A single val- cell or small cluster of val- cells is frequently observed at the center of this region (arrow in C). (D-F) krox20 staining (D,E) and mariposa staining (F) of transplant recipients. Brown cells are donor-derived (see Materials and Methods). (D) The distribution of wild-type cells in the hindbrain of a wild-type host. (E,F) The distribution of val- cells in a wild-type host, showing that mutant cells are specifically excluded from r5 and r6. The embryo shown in E is the same embryo as is shown in B and C, and the cell noted in C is observed to lie at the r5-6 boundary (arrow in E). In 31 out of 39 genetically mosaic embryos analyzed in which transplanted cells had contributed to the hindbrain, labelled val- cells were excluded from r5 and r6 although not necessarily from the boundary between them. In the remaining 8 mosaic embryos, val- cells were observed in r5 and/or r6, but these cells were located ventrally, either in or very near the floor plate (not shown). In contrast, 94 out of 100 control embryos into which labeled wild-type cells had been transplanted had labeled cells distributed throughout the hindbrain, including r5 and r6. The remaining 6 control embryos had relatively few transplanted cells and these tended to be localized to rhombomere boundaries. (G-J) A series of live images of rhodamine-labeled val- cells transplanted into a single wild-type host, shown in dorsal view. The time-points shown are 7 somites (12 h; G), 10 somites (14 h; H), 14 somites (16 h; I) and 24 h (J). As early as the 7-somite stage, val- cells begin to be excluded from the presumptive r5 and r6 of a wild-type host (indicated by a double arrow at the midline), with the exception of a few cells that ultimately lie at the r5-6 boundary (arrowheads). (K) Schematic diagram of the experimental approach used for L and M. Rhodamine-labeled cells from a wild-type donor were transplanted into a val- host embryo at the early gastrula stage. (L,M) Fluorescent and transmitted light images of the same horizontal section of a mutant host embryo. Anterior is to the left. Donor-derived cells in this experiment are fluorescently labelled. (L) Wild-type cells in a val- host form abnormal, unilateral clusters of rounded cells in rX, which is adjacent to the otic vesicle (ov), while they otherwise contribute normally to the brain and spinal cord. (M) The more anterior of these clusters, which lies at the r4-rX boundary, autonomously expresses krox20 although the surrounding mutant cells have failed to acquire r5 identity (arrow in L and M). Scale bars: B,C, 100 μm; D-F, 50 μm; G-J, 100 μm; L,M, 50 μm. |