- Title
-
Regulation of ddb2 expression in blind cavefish and zebrafish reveals plasticity in the control of sunlight-induced DNA damage repair
- Authors
- Zhao, H., Li, H., Du, J., Di Mauro, G., Lungu-Mitea, S., Geyer, N., Vallone, D., Bertolucci, C., Foulkes, N.S.
- Source
- Full text @ PLoS Genet.
|
|
|
|
|
|
|
|
|
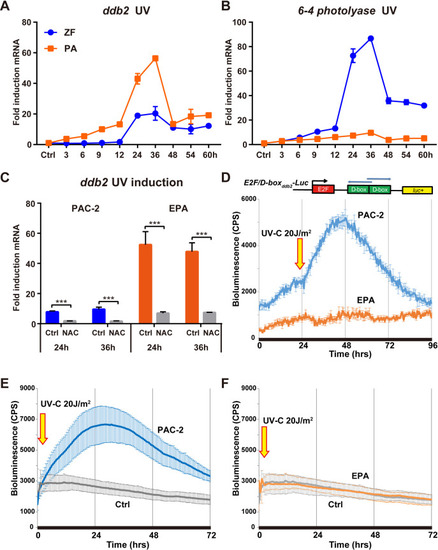
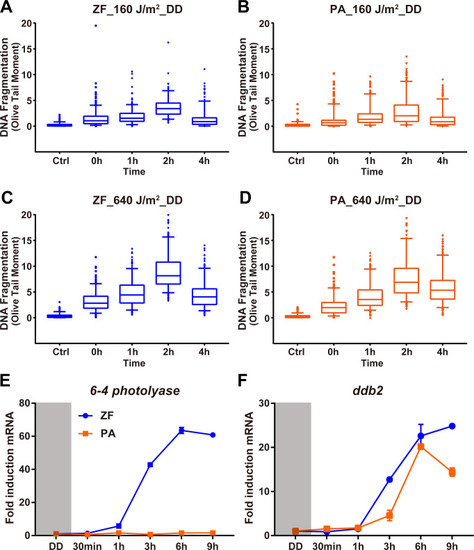
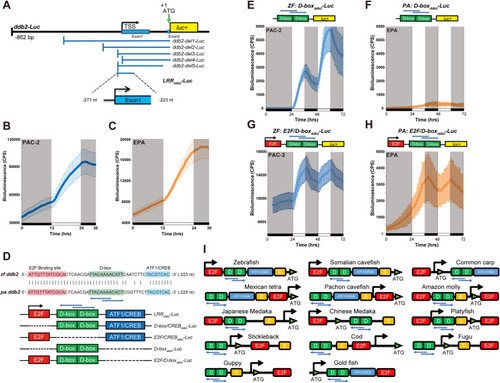
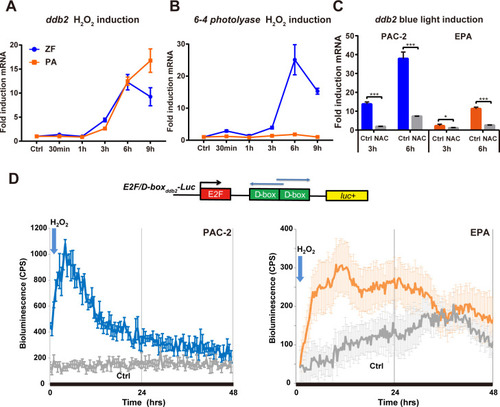
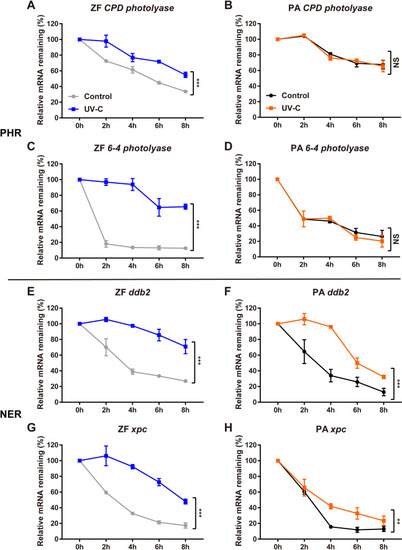
qRT-PCR analysis of |