- Title
-
A Conserved Notochord Enhancer Controls Pancreas Development in Vertebrates
- Authors
- Amorim, J.P., Gali-Macedo, A., Marcelino, H., Bordeira-Carriço, R., Naranjo, S., Rivero-Gil, S., Teixeira, J., Galhardo, M., Marques, J., Bessa, J.
- Source
- Full text @ Cell Rep.
|
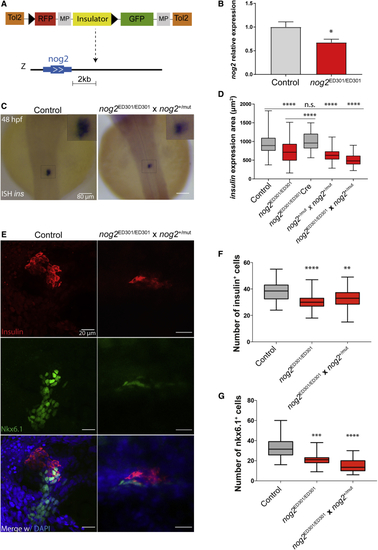
nog2 Is Required for Proper Pancreas Development (A) ED301 zebrafish line corresponds to an ED transposon integration containing a potent enhancer blocking insulator (yellow) and mapped 2 kb downstream of (B) In the ED301 zebrafish line, (C) (D) Quantification of the insulin expression area detected by (E) Representative confocal images of 48 hpf zebrafish embryos counterstained with a DAPI nuclear marker (blue), an anti-insulin antibody marking β cells (red), and an anti-Nkx6.1 antibody marking pancreatic progenitor cells (green). Images represent the maximum-intensity z projection of several focal planes obtained in a Leica Sp5 confocal microscope using a 40× objective. Scale bars represent 20 μm. (F) Quantification of the number of insulin-expressing cells in nog2ED301/ED301 and nog2ED301/ED301, nog2+/mut outcross embryos compared with controls (n ≥ 30). Error bars represent SD; ∗∗∗∗p < 0.0001, ∗∗p < 0.01. (G) Quantification of the number of nkx6.1-expressing cells in nog2 ED301/ED301 and nog2ED301/ED301, nog2+/mut outcross embryos compared with controls (n ≥ 18). Error bars represent SD; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001. |
|
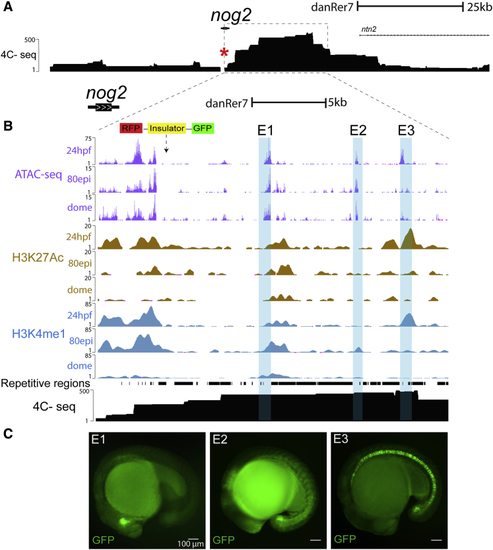
(A) Genomic landscape of (B) Zoom-in in the locus of (C) Representative images of GFP reporter lines for enhancers E1 to E3 (see also |
|
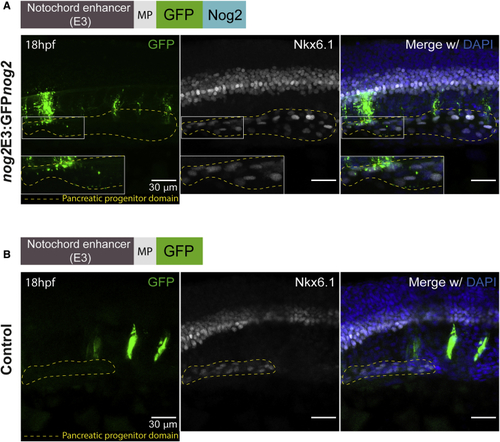
Nog2 Diffuses from the Notochord to the Pancreatic Progenitor Domain (A) Representative confocal image of a 18 hpf zebrafish embryo injected with the (B) Representative confocal image of a 18 hpf zebrafish embryo injected with nog2E3:GFP (control), in which the pattern of GFP expression (green) is restricted to the notochord. None of 18 analyzed embryos displayed colocalization of GFP aggregates with Nkx6.1-labeled cells. Embryos were counterstained with the nuclear marker DAPI (blue), and the endocrine progenitor domain is delimited by a yellow dashed line (see also EXPRESSION / LABELING:
PHENOTYPE:
|
|
The (A) Representation of the WT nog2E3 enhancer (above) and somatic deletions (below) generated by the injection of the Cas9 protein, together with sg1 and sg2 targeting two regions of the sequence 237 bp apart (see also (B) (C) Quantification of the (D) Diagram of the nog2E3:GFPnog2 construct used to achieve notochord-specific overexpression of GFPnog2 and the respective control. (E) (F) Quantification of the (G) Representative confocal images of 48 hpf zebrafish embryos counterstained with a DAPI nuclear marker (blue), an anti-insulin antibody marking β cells (red), and an anti-Nkx6.1 antibody marking pancreatic progenitor cells (green). Images represent the maximum-intensity z projection of several focal planes obtained in a Leica Sp5 confocal microscope using a 40× objective. Scale bars represent 20 μm. (H) Quantification of the number of insulin-expressing cells in nog2E3del1/del2 embryos compared with controls (n ≥ 19). Error bars represent SD; ∗p < 0.05. (I) Quantification of the number of nkx6.1-expressing cells in nog2E3del1/del2 embryos compared with controls (n ≥ 13). Error bars represent SD; ∗∗∗p < 0.001. |
|
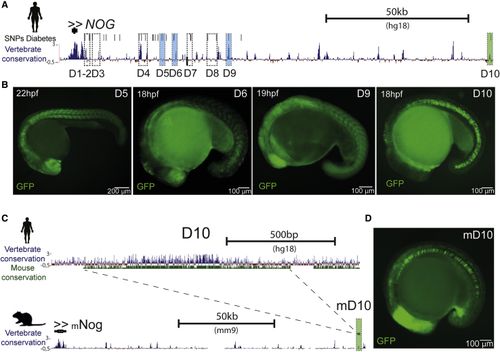
The Expression of (A) Genomic landscape of human (B) Representative images of GFP reporter lines for enhancers D5, D6, D9, and D10 (see also (C) Genomic landscape of the human D10 sequence, showing vertebrate (blue track) and mouse conservation (green track) (above). Genomic landscape of mouse (D) Representative image of the GFP reporter line for the mouse enhancer mD10. Scale bar represents 100 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
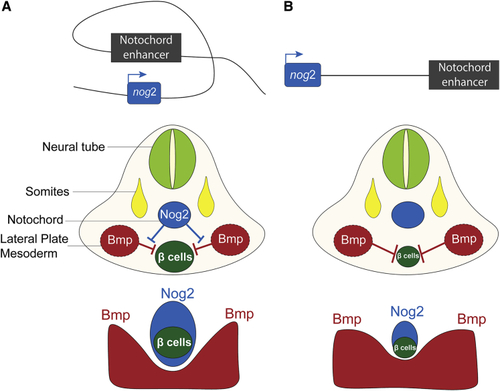
Proper Pancreas Development Requires the Expression of Nog2 in the Notochord (A) Diagram of the nog2 notochord enhancer driving expression of (B) When the activity of the |