- Title
-
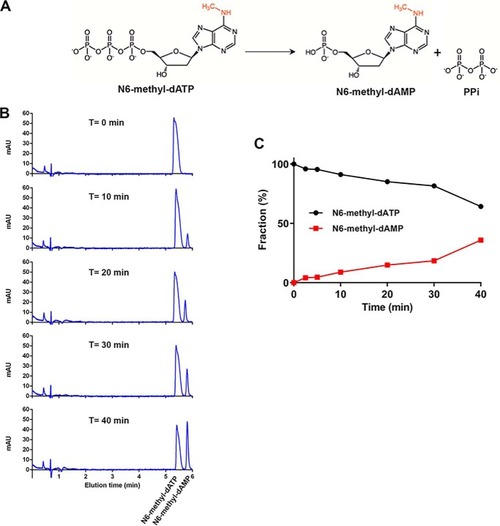
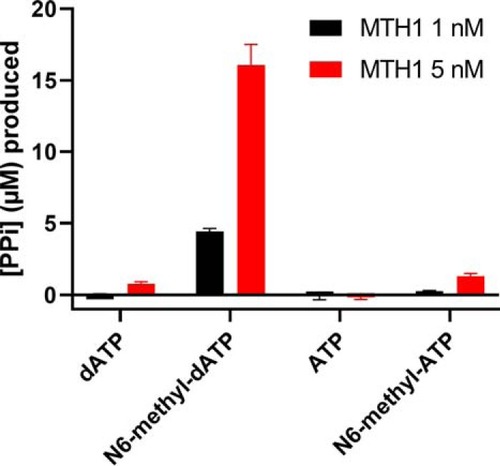
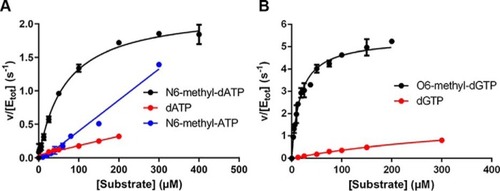
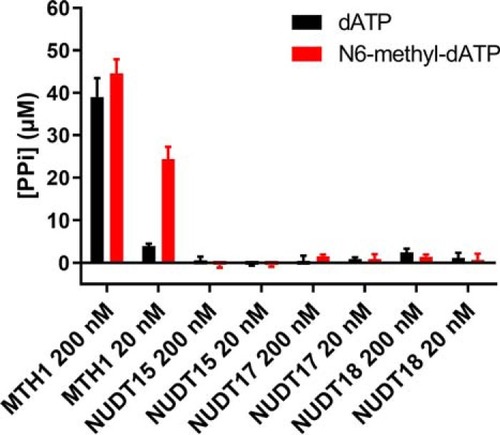
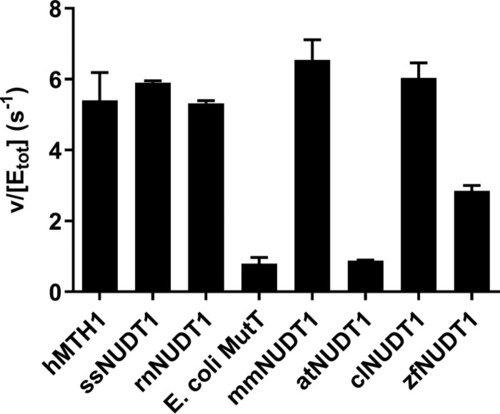
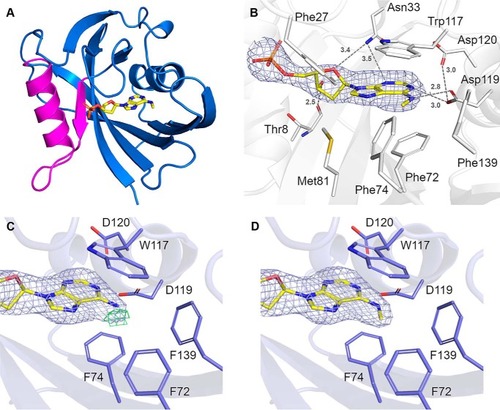
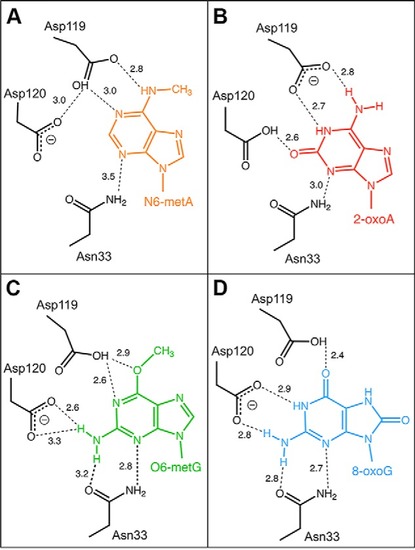
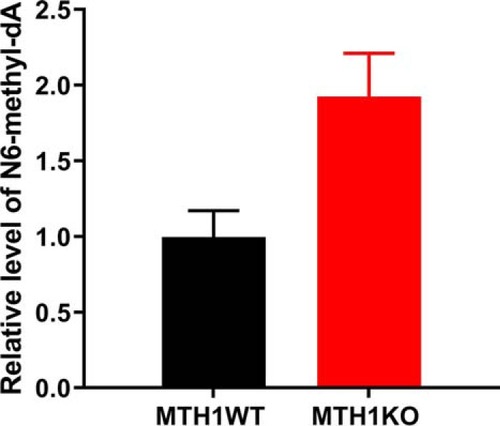
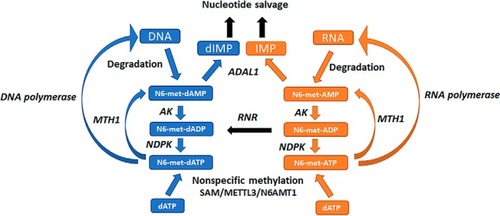
MutT homologue 1 (MTH1) removes N6-methyl-dATP from the dNTP pool
- Authors
- Scaletti, E.R., Vallin, K.S., Bräutigam, L., Sarno, A., Berglund, U.W., Helleday, T., Stenmark, P., Jemth, A.S.
- Source
- Full text @ J. Biol. Chem.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
PHENOTYPE:
|
|
|