- Title
-
Rigid Embedding of Fixed and Stained, Whole, Millimeter-Scale Specimens for Section-free 3D Histology by Micro-Computed Tomography
- Authors
- Lin, A.Y., Ding, Y., Vanselow, D.J., Katz, S.R., Yakovlev, M.A., Clark, D.P., Mandrell, D., Copper, J.E., van Rossum, D.B., Cheng, K.C.
- Source
- Full text @ J. Vis. Exp.
|
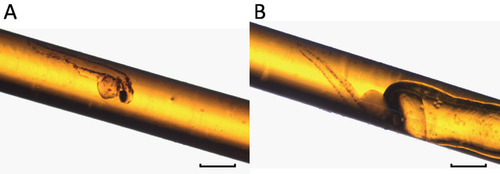
Successfully embedded specimens are devoid of air bubbles. (A) An embedded 3 day post-fertilization (dpf) larval zebrafish without air bubble. (B) An embedded 3 dpf zebrafish with an air bubble. Air trapped during the embedding process can move toward the specimen if the sample is not placed horizontally during the polymerization. Scale bars = 1 mm. |
|
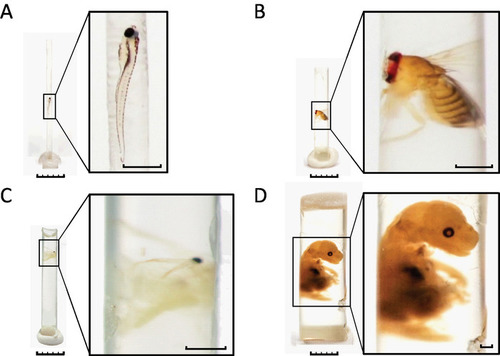
A wide variety of specimens can be embedded with our protocol. (A) 7 dpf zebrafish larva. (B) Adult Drosophila. (C) Adult Daphnia. (D) Mouse embryo. Larger samples such as the mouse embryo are accommodated by a few modifications to the protocol. Briefly, the polyimide tubing was filled to 1/3 of its length with resin and polymerized prior to embedding. Fixed and stained sample was placed on top of the pre-polymerized resin. The tubing was then filled with un-polymerized resin followed by a second polymerization. The polyimide tubing was removed prior to photography to allow better visualization of the sample in this figure. The removal of the tubing is not necessary for successful image acquisition by micro-CT. Scale bars: specimen images, 5 mm; enlarged insets, 1 mm. |
|
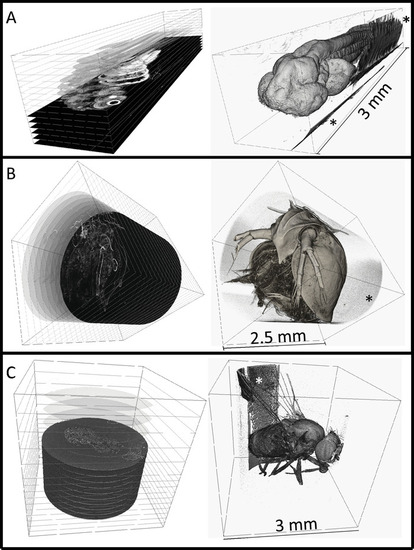
3-dimensional renderings of embedded specimens. (A) Reconstructed 3 dpf zebrafish larva at 0.743 × 0.743 × 0.743 µm3 voxel resolution. (B) Reconstructed adult Daphnia (3 × 3 × 3 µm3 voxel resolution). (C) Reconstructed adult Drosophila (3.1 × 3.1 × 3.1 µm3 voxel resolution). Digital cross-sections can be generated at any angle as shown on the left column. Surface renderings are shown on the right, rendered using commercial software. Embedding resin can be seen in all scans outside of the sample (noted by *), but does not interfere with the sample itself. The zebrafish larva was imaged at Argonne National Laboratory at the Advanced Photon Source that is synchrotron-based. The Daphnia or Drosophila specimens were imaged with a commercial X-ray microscope. |
|
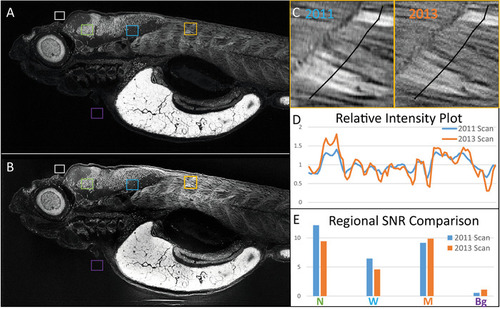
Samples embedded with our protocol can be re-imaged with micro-CT. The same 5 dpf zebrafish larva was imaged in (A) 2011 and (B) 2013, presented as average intensity projections of the sagittal view. Image comparison between scans after the storage shows the preservation of anatomical features. (C) A close-up of muscle from both scans is presented, with segmented lines indicating corresponding regions used for (D) intensity profile generation. Intensity profiles normalized to their corresponding averages along both lines show spatially matching local peaks in both scans. (E) Average intensity values for selected regions in A and B belonging to background (Bg, purple), neuronal nuclei (N, green), white matter (W, blue), and the muscle (M, orange) were divided by an average for background (white) and used to generate signal-to-noise ratios (SNR), which corresponds well between scans. Scale bars = 100 µm. |