- Title
-
Kremen1 restricts Dkk activity during posterior lateral line development in zebrafish
- Authors
- McGraw, H.F., Culbertson, M.D., Nechiporuk, A.V.
- Source
- Full text @ Development
|
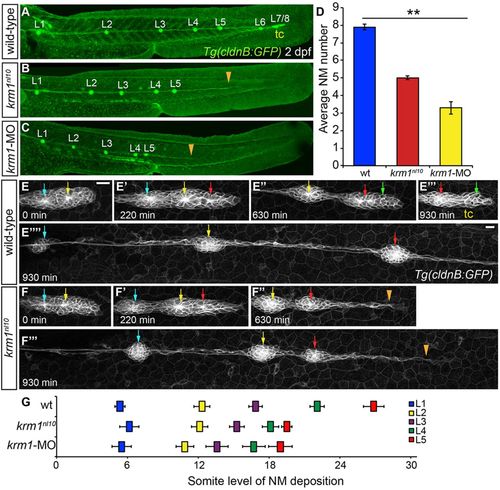
The pLL is truncated in krm1nl10 mutants. (A-C) Confocal projections of 2dpf larvae expressing Tg(cldnB:GFP), (L1-L8: pLL NMs 1-8). (A) WT embryo showing completed pLLP migration and deposition of all NMs. (B) In a krm1nl10 mutant, there are fewer deposited NMs and the pLLP stalled prematurely (orange arrowhead). (C) kremen1 morphants show a similar reduction of NMs and pLLP stalling (orange arrowhead). (D) Quantification of neuromasts in WT, krm1nl10 mutant and krm1-MO-injected embryos (n=10 embryos per condition; **P<0.001, one-way ANOVA). (E-F4) Still confocal projections from live 930-min time-lapse movies showing pLLP migration in WT and krm1nl10 embryos beginning at 36hpf. At 0min, WT and mutant primordia contain two proto-NMs (E,F, blue and yellow arrows). By 220min, both WT and mutant primordia have added an additional proto-NM (E2,F2, red arrow). (E3) At 630min, the WT pLLP contains the proto-NMs that will form the terminal cluster (green arrow). By contrast, the krm1nl10 pLLP fail to form a new proto-NM and migrate as a thin trail of cells (F3, orange arrowhead). By 930min, WT pLLP has migrated out of frame (EW) and formed the tc (E4). The mutant pLLP has stalled (a thin trail of cells extended from the last deposited NM, marked by orange arrowhead; F4). (G) Quantification of L1-L5 NM location based on somite level (tc is not included). NMs are shifted anteriorly in krm1nl10 mutant and krm1-MOrphants relative to WT embryos (n=6 embryos per condition; P<0.001, two-way ANOVA with replication). Scale bars: 20μm. NM, neuromast; tc, terminal cluster. |
|
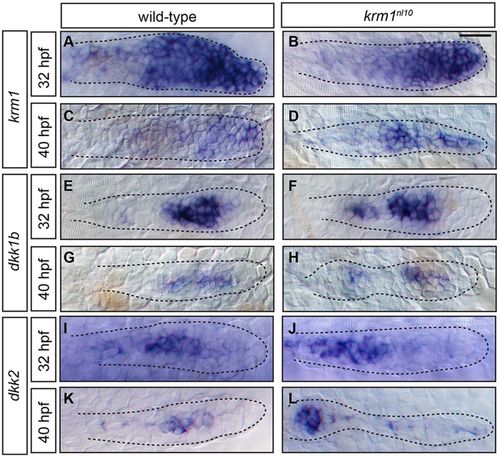
kremen1, dkk1b and dkk2 are expressed in the pLLP of WT and krm1nl10 mutant embryos. (A-L) Expression of kremen1, dkk1b and dkk2 in the primordia (highlighted by dashed lines) of WT and krm1nl10 embryos at 32 and 40hpf. kremen1 expression is progressively decreased and restricted to the leading zone between 32 and 40hpf in WT (A,C) and krm1nl10 (B,D) embryos. dkk1b is expressed in the mid-zone of the pLLP at 32 and 40hpf in WT (E,G) and krm1nl10 (F,H) embryos. dkk2 is expressed in the mid-zone at 32hpf in WT (I) and mutant (J) primordia. At 40hpf, dkk2 is expressed in the mid-zone of WT pLLP (K) and in the last deposited NM in mutant embryos; faint expression is present throughout the mutant pLLP (L). Scale bar: 20μm. |
|
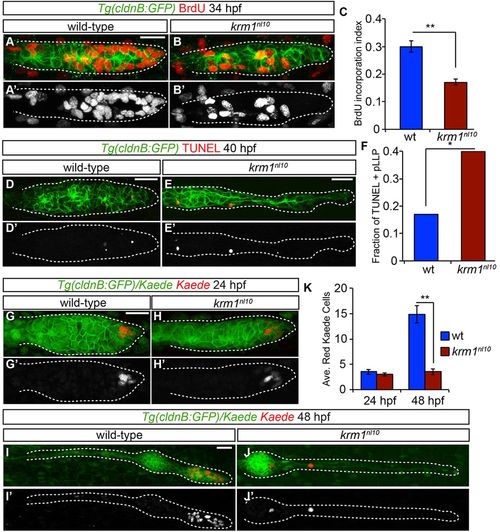
Cellular proliferation and survival are impaired in krm1nl10 pLLP. (A-B2) BrdU incorporation in WT and krm1nl10 primordia between 32.5 and 34hpf. (C) Quantification of cells that have incorporated BrdU, showing a significant decrease in krm1nl10 mutant embryos versus WT (n=10 WT and 9 mutant embryos; **P<0.001, Student′s t-test). (D-E2) Confocal projections of pLLP at 40hpf labeled with TUNEL (red) in Tg(cldnB:GFP)+ (green) in WT (D,D2) and krm1nl10 (E,E2) embryos. (F) TUNEL-positive fractions of pLLPs. Significantly more krm1nl10 mutant pLLPs contained TUNEL-positive puncta (n=35 WT and 40 mutant embryos; *P<0.03, Fisher′s Exact Test). (G-J2) Confocal projections of live Tg(cldnB:GFP) (green) embryos at 24hpf labeled with nuclear-localized Kaede before (green) and after (red) photoconversion. At 24hpf, both WT (G,G2) and krm1nl10 (H,H2) embryos contain photoconverted Kaede cells in the leading zone of the pLLP. At 48hpf, photoconverted Kaede cells in the WT embryo have divided and are localized in the tc NMs (I,I2). In the krm1nl10 embryo, photoconverted cells have not proliferated (J,J2). (K) Quantification of photoconverted Kaede cells at 24 and 48hpf; there are significantly fewer converted cells at 48hpf in krm1nl10 mutant pLLP compared with WT (n=24 WT and 15 krm1nl10 mutant embryos; **P<0.001, Student′s t-test). Scale bars: 20μm. |
|
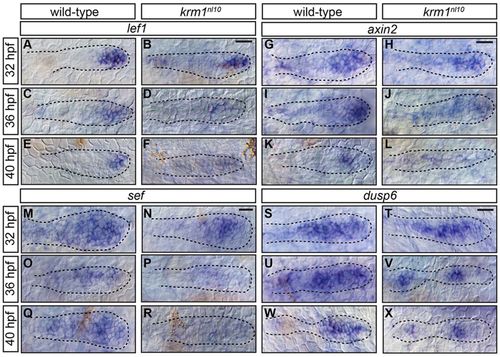
Wnt activity is prematurely and progressively decreased in krm1nl10 mutants. (A-R) Expression of the Wnt targets, lef1, axin2 and sef, in the pLLP at 32, 36 and 40hpf in WT and krm1nl10 embryos. In WT primordia, expression domains of lef1 (A,C,E), axin2 (G,I,K) and sef (M,O,Q) are progressively decreased between 32 and 40hpf. In krm1nl10 mutant primordia, Wnt target expression is similar to WT at 32hpf (B,H,N). At 36hpf, expression domains are decreased in mutants compared with WT (D,J,P), and are absent in mutants at 40hpf (F,L,R). dusp6 is expressed in the leading and mid-zones of WT primordia between 32 and 40hpf (S,U,W); expression is absent in the leading zone of krm1nl10 mutant primordia at these stages (T,V,X). Scale bars: 20 μm. |
|
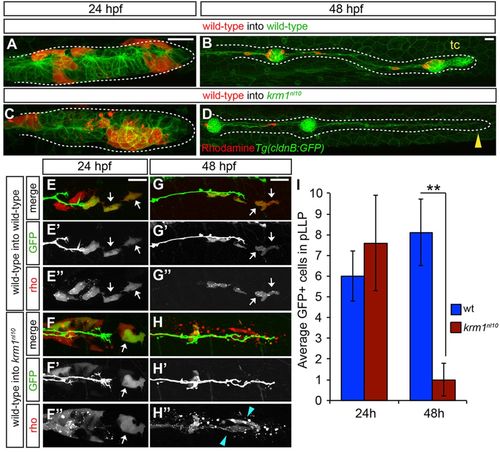
Loss of Kremen1 leads to non-cell-autonomous defects in the pLLP. (A-D) Live confocal projections of mosaic, Tg(cldnB:GFP) WT or krm1nl10 embryos containing rhodamine-labeled donor cells (red) from WT donor embryos at 24 and 48hpf. At 24hpf, donor cells are found in the leading region of pLLP in WT (A) and mutant (C) hosts. At 48hpf, the pLLP shown in A has migrated along the length of the trunk and formed the terminal cluster (tc; B), whereas the mutant pLLP in C has failed to migrate and diminished to a thin trail of cells (D, yellow arrowhead). (E-H3) Live confocal projections of WT and krm1nl10 host embryos that express TgBAC(neuroD:eGFP) and contain donor cells from Tg(7xtcf-siam:eGFP) donor embryos labeled with rhodamine (red). At 24hpf, the leading region of primordia of both WT (E-E3) and mutant (F-F3) hosts contain GFP-positive (E2,F2) and rhodamine-positive (E3,F3) donor cells (white arrows). At 48hpf, GFP/rhodamine-positive cells remain in the leading zone of WT primordia (G-H3, white arrows). In krm1nl10 mutant primordia, few GFP-positive cells remain (H-H2), and the remaining rhodamine-positive donor cells are fragmented and appear to undergo cell death (H3 blue arrowheads). (I) Quantification of Tg(7xtcf-siam:eGFP)-positive donor cells at 24 hpf and 48 hpf in control (WT donor cells in WT hosts) and experimental (WT donor cells in krm1nl10 hosts) primordia (n=5 krm1nl10 hosts, 10 WT hosts; **P=0.007, Student′s t-test). Scale bars: 20μm. |
|
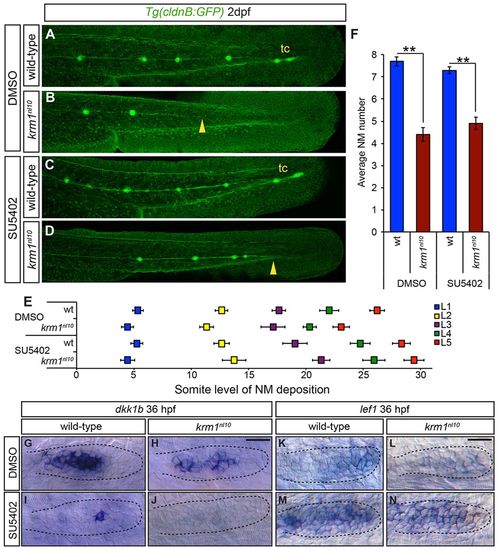
Inhibition of Fgf signaling partially rescues NM spacing in krm1nl10 mutants. (A-D) Confocal projection of 2dpf Tg(cldnB:GFP) WT or krm1nl10 embryos, treated with either DMSO (controls) or SU5402. (A,E) DMSO-treated WT embryos show full pLLP extension and tc formation. (B,E) krm1nl10 mutants treated with DMSO show pLL truncation midway along the trunk (yellow arrowhead). (C,E) Suboptimal SU5402 treatment of WT embryos results in normal pLL extension and tc formation. (D,E) In krm1nl10 mutants, pLLP migration is partially rescued and pLL is extended to the end of the tail (yellow arrowhead), although tc NMs are rarely present (n=10 embryos per condition; P<0.001, two-way ANOVA with replication). (F) NM numbers are not significantly different between WT embryos treated with DMSO or SU5402 and between krm1nl10 mutants treated with DMSO or SU5402 (n=10 embryos/condition; **P<0.001, Student′s t-test). (G-N) Expression of dkk1b or lef1 in WT and krm1nl10 pLLPs treated with DMSO or SU5402 at 36 hpf. dkk1b is expressed in the mid-region of the pLLP of DMSO-treated WT (G) and mutant (H) embryos. In SU5402-treated embryos, dkk1b expression is reduced in WT pLLP (I) and absent in krm1nl10 pLLP (J). lef1 is expressed in the leading region of WT primordia (K) and absent in krm1nl10 mutant primordia treated with DMSO (L). SU5402 treatment results in increased lef1 expression in WT (M) and mutant (N) primordia. Scale bars: 20μm. |
|
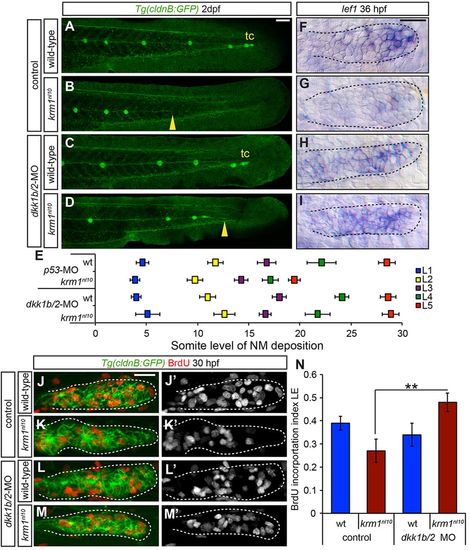
dkk1b/2 morpholino injections rescue krm1nl10 pLL formation. (A-D) Confocal projection of 2dpf Tg(cldnB:GFP) WT or krm1nl10 embryos, injected with either control or dkk1b/2-MOs. (A) Control WT embryos show full pLLP migration and tc formation. (B) Control krm1nl10 embryos display characteristic premature stalling of the pLLP (yellow arrowhead). (C) Injection of dkk1b/2-MOs in WT embryos results in full pLLP migration and tc formation, although NM spacing is shifted posteriorly. (D) In krm1nl10 mutants injected with dkk1b/2-MOs, pLLP migration and pLL formation are partially rescued (yellow arrowhead). (E) NM spacing in krm1nl10 mutants injected with dkk1b/2-MOs is not significantly different from WT control NMs (n=8 embryos/condition; P<0.001, two-way ANOVA with replication). (F-I) lef1 expression at 36 hpf is restricted to the pLLP leading region in control WT pLLP (F) and downregulated in control krm1nl10 mutant pLLP (G). dkk1b/2-MO injection results in an expansion of lef1 expression in both WT (H) and krm1nl10 (I) primordia. (J-M2) Confocal projections of BrdU incorporation in both WT and krm1nl10 control and dkk1b/2-MO-injected embryos at 30 hpf. BrdU incorporation index is high in the leading region of control WT embryos (J,J2,N) and significantly decreased in krm1nl10 mutants (K,K2,N). dkk1b/2-MO injection results in no change in WT (L,L2,N) and in a significant increase in the BrdU incorporation index in the leading region of krm1nl10 mutant primordia (M,M2,N) (n=10 embryos per condition; **P=0.007, Student′s t-test). Scale bars: 20 μm. |
|
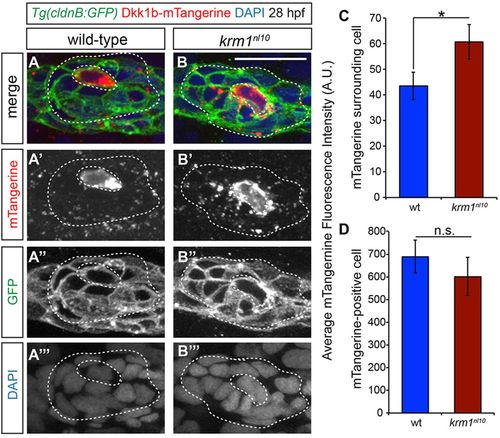
Mosaic expression of Dkk1b-mTangerine shows a greater spread in krm1nl10 primordia. (A-B4) Confocal projections of 28hpf WT and krm1nl10 pLLP showing mosaic expression of Dkk1b-mTangerine driven by heat shock promoter, which was activated at 25hpf. (A-A4) WT showing expression of Dkk1b-mTangerine in a single cell (inner dashed lines) and low expression levels in surrounding cells (outer dashed lines). (B-B4) krm1nl10 mutant expressing Dkk1b-mTangerine in a single cell (inner dashed lines) and punctate expression in surrounding cells (outer dashed lines). (C) Quantification of mTangerine fluorescence intensity in arbitrary units (A.U.). ROI: two cell diameters from the mTangerine-expressing source cell, excluding the source cell, minus background. (D) Quantification of mTangerine expression intensity in the source cell (A.U.) shows no significant difference between WT and mutant cells. Note that krm1nl10 mutants show a significant increase in fluorescence intensity compared with WT (n=31 WT and n=30 krm1nl10 primordia in six separate experiments; **P<0.04, Student′s t-test). Scale bar: 20 μm. |
|
The pLL is truncated in krm1nl10 adults (A,B) DASPEI labeling of 4 months post fertilization (mpf) Tg(cldnB:GFP) expressing wild-type and krm1nl10 mutant adults. (A) A wild-type adult (24 mm in length) with DAPSEI labeled NMs on the trunk (blue arrows) and tail (blue brackets). (B) A 21 mm krm1nl10 mutant adult lacks the posterior-most trunk NMs (blue arrows) and all tail NMs. PHENOTYPE:
|
|
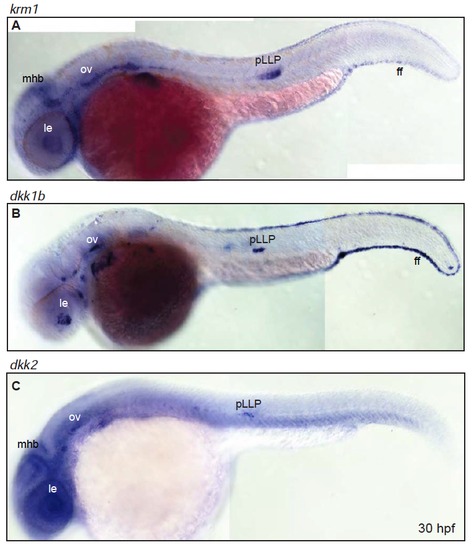
Expression of krm1, dkk1b and dkk2 partially overlaps throughout the embryo (A-B) Expression domains of krm1 and its ligands dkk1b and dkk2 in 30 hpf wildtype embryos. (A) krm1 is expressed in the mid-hindbrain boundary (mhb), lens (le), otic vesicle (ov), posterior lateral line primordium (pLLP) and the fin fold (ff). (B) dkk1b is expressed in the le, ov, pLLP and ff. (C) dkk2 is expressed in the mhb, le, ov and pLLP. EXPRESSION / LABELING:
|
|
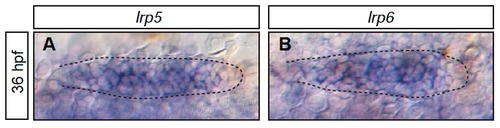
The Wnt receptors lrp5 and lrp6 are expressed in the pLLP (A-B) Expression of lrp5 (A) and lrp6 (B) throughout the primordia at 36 hpf in wild-type embryos. Scale bars=20 μm. EXPRESSION / LABELING:
|
|
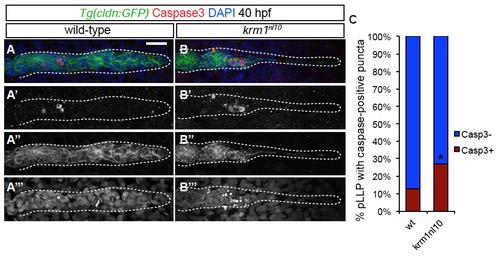
Cell death is increased in krm1nl10 mutant primordia (A-B′′′) Confocal projections of 40 hpf wild-type (A-A′′′) and krm1nl10 (B-B′′′) labeled with activated Caspase3 antibody. Scale bars=20 μm. (n=35 embryos per condition, *p<0.01, Fisher′s Exact Test). |
|
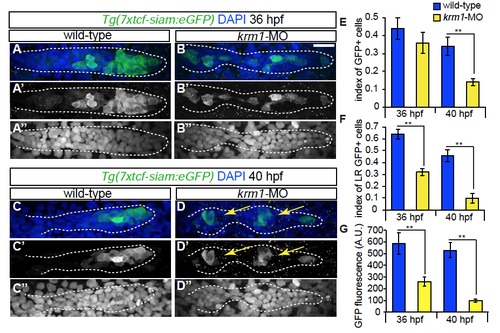
Wnt activity is decreased following Krm1 knockdown. (A-D′′′) Confocal projections of wild-type and kremen1 morphant embryos expressing the Wnt sensor Tg(7xtcf-siam:eGFP) (green); nuclei are labeled with DAPI (blue). (A-A′′, C-C′′) GFP is expressed in cells of the leading part of a wildtype pLLP at 36 and 40 hpf. (B-B′′,D-D′′) In kremen1-MO injected embryos, GFP-positive cells are located throughout the pLLP, most are incorporated into proto-NMs (yellow arrows) and notably reduced in the leading region. By 40 hpf, the index of GFP-positive cells is significantly reduced in kremen1 morphants (E). (F) The index of GFP in the leading zone of the pLLP (the caudal-most 30 cells of the pLLP) is significantly reduced in kremen1 morphants at 36 and 40 hpf as compared to wild-type controls (n=10-12 embryos/condition; **p<0.001 Student’s t-test). Scale bars=20μm. (G) The mean fluorescence intensity of GFP-positive cells, measured in arbitrary units (A.U.), was significantly reduced in morphant primordia at both 36 and 40 hpf (n=10-12 embryos/condition; **p<0.001 Student’s t-test). EXPRESSION / LABELING:
|
|
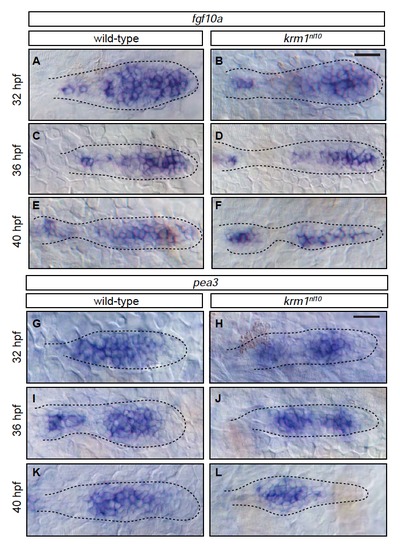
Members of the Fgf pathway are expressed in wild-type and krm1nl10 primordia(A-F) Expression of fgf10a in wild-type and krm1nl10 primordia between 32 and 40 hpf. In wild-type embryos, fgf10a is expressed in the leading region of the primordia at 32, 36 and 40 hpf (A,C,E). A similar expression pattern for fgf10a was seen in krm1nl10 primordia (B,D,F). (G-L) Expression of the Fgf target pea3 between 32 and 40 hpf. In wild-type (G,I,K) and krm1nl10 (H,J,L) embryos, expression of pea3 is seen in the mid-zone of the primordia. Scale bars=20 μm. |
|
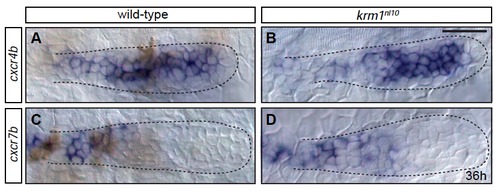
Expression domains of chemokine receptors are not altered in krm1nl10 mutants (A-D) Expression of cxcr4b and cxcr7b at 36 hpf in the primordia of wild-type and krm1nl10 mutants. cxcr4b is expressed in the leading and mid-zones of wild-type (A) and krm1nl10 (B) primordia. cxcr7b is expressed in the trailing zone of the wild-type (C) and mutant (D) primordia. Scale bars=20 μm. EXPRESSION / LABELING:
|
|
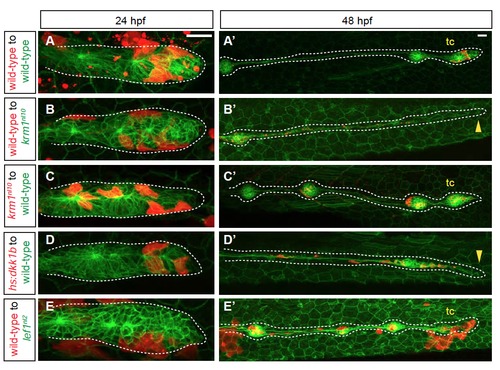
pLL formation in mosaic embryos that contain krm1nl10, Tg(hsp701:dkk1b-GFP) and lef1nl2 cells (A-E′) Confocal projections of chimeric embryos with rhodamine-labeled donor cells (red) in Tg(cldnB:GFP)-positive hosts. At 24 hpf, donor cells are present in the leading region of primordia in wild-type to wild-type chimeras (A), wild-type to krm1nl10 (B), krm1nl10 to wild-type (C), Tg(hsp701:dkk1b-GFP) to wild-type (D) and wild-type to lef1nl2 mutants (E). At 2 dpf, pLL extension and terminal cluster (tc) formation was seen in wild-type to wild-type (A′), krm1nl10 to wild-type (C’)and wild-type to lef1nl2 (E′). pLL formation was truncated in wild-type to krm1nl10 (B′) and Tg(hsp701:dkk1b-GFP) to wild-type (D′) chimeras. Scale bars=20 μm. |
|
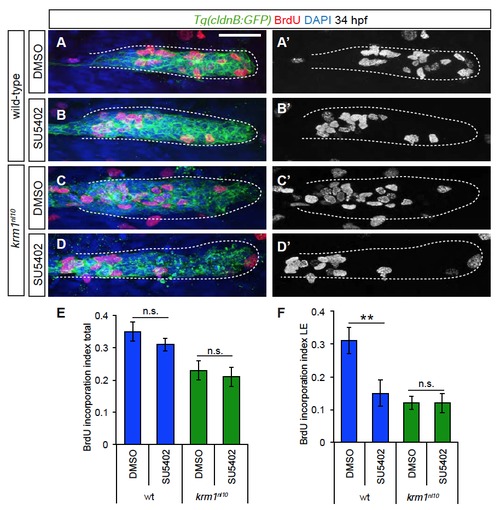
Cellular proliferation is not rescued by treatment with SU5402 (A-D′) Confocal projections of BrdU incorporation (red) in wild-type and krm1n10 embryos treated with DMSO or SU5402 between 24 and 34 hpf. BrdU label is present in the leading pLLP cells in DMSO treated wild-type (A-A′), but not mutant embryos (C-C′). SU5402 treatment resulted in reduced leading region BrdU incorporation in wild-type embryos (B-B′), but incorporation was not altered in SU5402 treated krm1nl10 mutants (D-D′). Scale bars=20 μm. (E) Quantification of the index of total BrdU incorporation in the pLLP showing no significant change in control versus treated wild-type embryos or control versus treated krm1nl10 mutants (n=11 embryos per condition, Student′s t-test). (E) Quantification of the index of BrdU incorporation in the leading region of the pLLP shows a significant decrease in SU5402 treated wild-type embryos, but no change in krm1nl10 mutants (n=11 embryos per condition, Student′s t-test). |
|
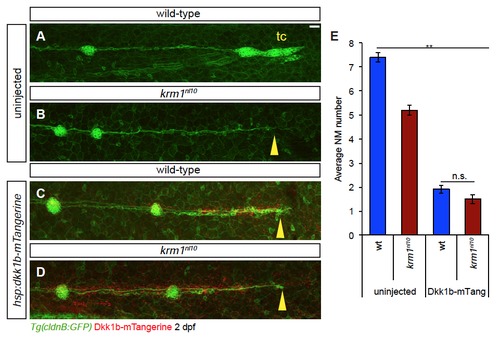
Ectopic expression of Dkk1b-mTangerine results in decreased NM number and pLL truncation (A-D) Confocal projections of 2 dpf embryos with ectopic expression Dkk1bmTangerine (red) driven by heat-shock at 25 hpf. (A) Uninjected wild-type embryo show full pLL extension and terminal cluster (tc) formation, while uninjected krm1nl10 mutants show pLL truncation (B; yellow arrowhead). Dkk1bmTangerine expression results in truncated pLL extension in both wild-type (C; yellow arrowhead) and krm1nl10 (D; yellow arrowhead) embryos. Scale bars=20 μm. (E) Quantification of NM number at 2 dpf; there is a significant decrease in NM between uninjected control embryos (wild-type and krm1nl10 mutants) and embryos expressing Dkk1b-mTangerine (n=15, **p<0.001, two-way ANOVA with replication). There was no difference in NM number between wild-type and mutants expressing Dkk1b-mTangerine (n=15; Tukey’s Post-Hoc Test). |