- Title
-
Function and Regulation of AUTS2, a Gene Implicated in Autism and Human Evolution
- Authors
- Oksenberg, N., Stevison, L., Wall, J.D., and Ahituv, N.
- Source
- Full text @ PLoS Genet.
|
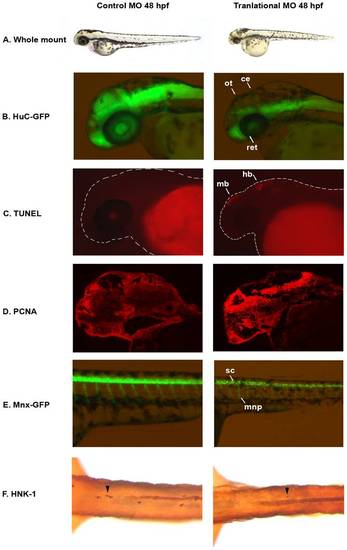
auts2 48 hpf morphant phenotype. (A) Fish injected with the 5 base-pair translational MO mismatch control have similar morphology as wild type fish. Injection of the auts2 translational MO results in fish with a stunted development phenotype that includes a smaller head, eyes, body and fins. (B) HuC-GFP fish injected with the 5 bp control MO display normal levels of developing neurons in the brain. HuC-GFP translational MO injected fish display considerably less developing neurons in the optic tectum (ot), retina (ret), and cerebellum (ce). (C) 5 bp mismatch control injected fish have little to non-observable apoptosis in the brain as observed by TUNEL staining, while translational MO injected fish display high levels of apoptosis, primarily in the midbrain (mb) and hindbrain (hb). (D) PCNA cell proliferation assay in the 5 bp MO control injected fish shows lower levels of cell proliferation in the brain compared to the translational MO injected fish. (E) Tg(mnx1:GFP) fish injected with the 5 bp MO control display normal levels of motor neurons versus the auts2 translational MO injected fish which have fewer motor neurons in the spinal cord (sc). In addition, motor neuron projections (mnp) are weaker and more perpendicular to the spinal cord. (F) Translational MO injected fish display fewer Rohon-Beard cells (arrowheads) in the spinal cord than morphants. All morphant fish are scaled to their 5 bp control counterparts. PHENOTYPE:
|
|
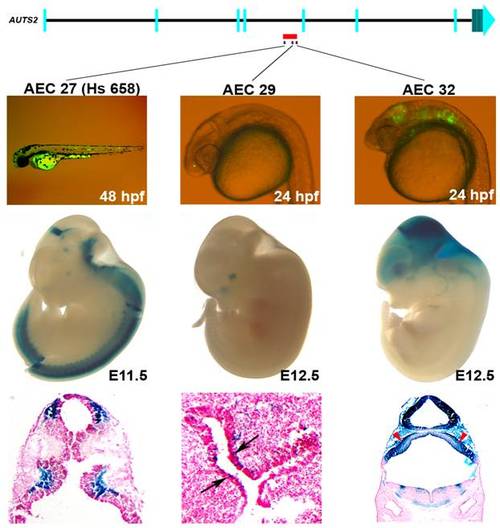
Enhancers within an ASD–associated AUTS2 intronic deletion [5]. Three positive enhancers (AEC27, 29, 32) show positive enhancer activity in zebrafish (24 or 48 hpf) and in mice (E11.5 or 12.5). AEC27 shows enhancer expression in the somitic muscle in zebrafish, while in mouse at E11.5 (hs658; [34]) it is active in the midbrain, medulla, and neural tube at E11.5. The histological section below shows its enhancer activity in the pretectum and the pons. At E12.5, AEC29 shows enhancer activity in the olfactory epithelium (arrows in histological section) similar to zebrafish and in addition also displays enhancer expression in the eye. AEC32 recapitulates the zebrafish enhancer expression displaying strong enhancer activity in the midbrain (tectum) and hindbrain and in addition also displays enhancer expression in the forebrain at E12.5. Histological sections of AEC32 show enhancer activity in the mouse cerebellum (red arrowheads). |
|
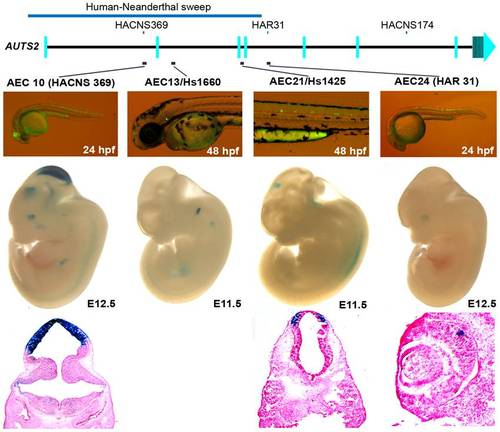
Four positive zebrafish and mouse enhancers in regions implicated in human evolution. At E12.5, AEC10 shows zebrafish and mouse enhancer expression in the midbrain and eye. The histological section below highlights its expression in the tectum. AEC13, is expressed in the otic vesicle both in zebrafish and E11.5 mouse embryos (hs1660 ; [34]). AEC21 is expressed in the spinal cord in zebrafish, while in the mouse it showed midbrain expression at E11.5 (hs1425; [34]). Histological sections below show its expression in the pretectum of the midbrain. AEC24 was expressed in the spinal cord and hindbrain in zebrafish and in the eye in mouse at E12.5. |
|
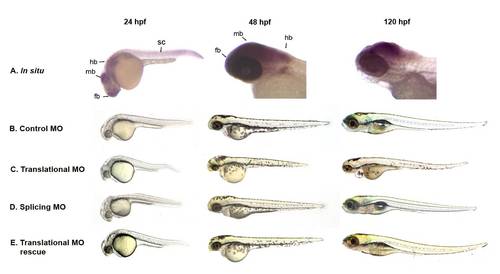
auts2 expression and morphant phenotype. (A) Whole-mount in situ hybridization of auts2 shows that it is expressed in the forebrain (fb) (including olfactory organs), midbrain (mb), hindbrain (hb), spinal cord (sc), the caudal peduncle and eye at 24hpf. At 48hpf, auts2 is expressed in the brain, pectoral fin and eye. At 120 hpf expression is restricted to the brain, primarily the midbrain, and weakly in the eye. (B) Fish injected with the 5 bp translational MO mismatch control have indistinguishable morphology as wild type fish at 24, 48 and 120 hpf. (C) Injection of the auts2 translational MO results in fish with a stunted development phenotype that includes smaller heads, eyes, bodies and fins. (E) Injection of the auts2 splice-blocking MO shows a similar but less severe phenotype than the auts2 translational MO. (E) The auts2 translational MO phenotype is partially rescued by co-injecting the full length human AUTS2. Note the longer body and larger brain compared to the translational and splicing morphant fish. MO injected fish in C, D, and E are scaled to the 5 bp injected control fish in B. |
|
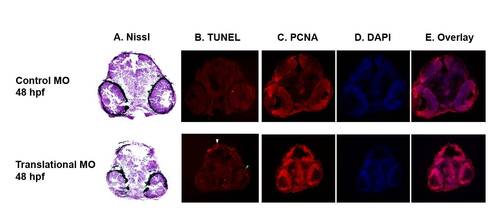
Histological phenotype of auts2 morphants (A) Nissl staining shows a reduction in neuron territory, primarily in the midbrain, of fish injected with the translational MO compared to 5 bp mismatch controls at 48 hpf. (B) TUNEL stained sections show fewer apoptotic cells in the optic tectum (white arrowhead) and the retina (green arrowhead) in 48 hpf auts2 morphants versus the 5 bp translational MO mismatch control. (C–E) Coronal sections stained with PCNA, DAPI and overlays show an increase in cell proliferation in the translational morphant fish compared to the 5 bp mismatch control in the mesencephalon, diencephalon and retina. PHENOTYPE:
|
|
znp-1 antibody on control and morphant fish. The motor neurons axons of the morphant fish are different than the controls, signified by a drastic increase in the amount of branching (red arrow). PHENOTYPE:
|
|
AUTS2 enhancer candidates (AECs) positive for enhancer activity in zebrafish. A representative fish of each positive AEC enhancer is shown. The number in the top right of every image is the AEC number and the hours post fertilization (hpf) when the picture was taken is indicated in the bottom right. Their tissue-specific expression pattern is denoted in Table S1 and http://zen.ucsf.edu. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|