- Title
-
Illuminating cell-cycle progression in the developing zebrafish embryo
- Authors
- Sugiyama, M., Sakaue-Sawano, A., Iimura, T., Fukami, K., Kitaguchi, T., Kawakami, K., Okamoto, H., Higashijima, S.I., and Miyawaki, A.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
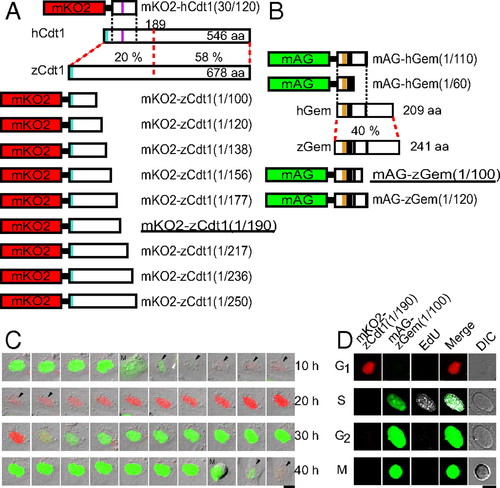
Development of fluorescent indicators for cell-cycle progression in fish cells (zFucci), and characterization of zFucci in transgenic fish (Cecyil) cells compared with Fucci in HeLa cells. (A) Structural domains of hCdt1 (human Cdt1) and zCdt1 (zebrafish Cdt1). Cyan box, PIP box or QXRVTDF motif (amino acids 3–9); violet box, Cy motif (amino acids 68–70). The N-terminal 189 aa of hCdt1 are sufficient for G1-specific accumulation of the protein (11). This regulatory region shows 20% sequence homology between hCdt1 and zCdt1. The other region, which contains the geminin and MCM6 binding domains, shows 58% homology between the two proteins. The G1 marker of the original Fucci, mKO2-hCdt1(30/120), is illustrated at the Top. Various constructs with concatenated mKO2 and deletion mutants of zCdt1 are shown at the Bottom. mKO2-zCdt1(1/190) is underlined. (B) Structural domains of hGem (human geminin) and zGem (zebrafish geminin). The S/G2/M markers of the original Fucci, mAG-hGem(1/110) and mAG-hGem(1/60), are illustrated at the Top. Two constructs with concatenated mAG and deletion mutants of hGem for labeling nuclei in S, G2, and M phases. mAG-zGem(1/100) is underlined. Orange box, D (destruction) box; black box, NLS (nuclear localization signal). (C) Cell-cycle-dependent changes in fluorescence of zFucci (mKO2-zCdt1(1/190) and mAG-zGem(1/100)) in Cecyil cells. Arrows indicate cells that were tracked. (Scale bar, 10 μm.) M, M phase. (D) Typical fluorescence images of Cecyil cells expressing zFucci (mKO2-zCdt1(1/190) and mAG-zGem(1/100)) and fluorescence from incorporated EdU (white) at G1, S, G2, and M phases. (Scale bar, 10 μm.) |
|
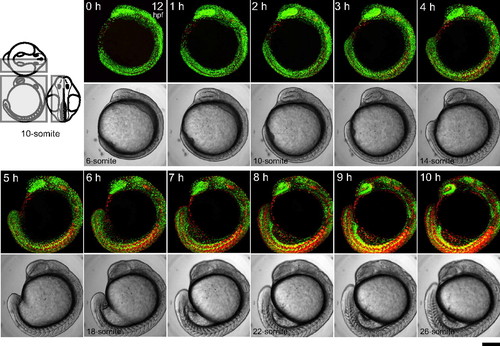
Time-lapse imaging of a Cecyil embryo during segmentation. Three-dimensional time-lapse imaging was performed to collect fluorescence images from the left half of the embryo at 10-min intervals using an Olympus FV1000 upright confocal microscope equipped with an objective lens (x10 N.A. 0.3). A dechorionated embryo in the early segmentation period at 12 hpf (6-somite stage) was mounted with the left side up in a chamber containing 0.3% agar. Since the sample was kept at less than 28 °C during observation, developmental stages cannot be accurately expressed in hpf. Due to z-stacking, green and orange signals at different z-positions merge to generate yellow signal. Note that zFucci does not yield yellow fluorescence at the G1/S transition, whereas the original Fucci in mammalian cells does. The scanned region is indicated by the gray box on the three views of an embryo at the 10-somite stage. (Scale bar, 200 μm.) |
|
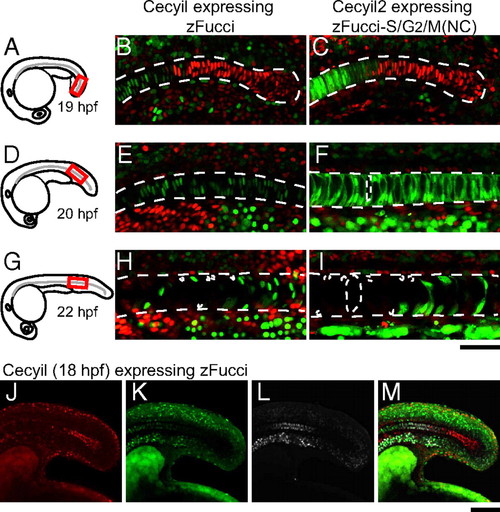
Cell-cycle transition waves in the differentiating notochord. Dechorionated embryos at various stages were fixed in 4% paraformaldehyde (PFA) solution, then each sample was mounted in 0.3% agar so that a confocal image of the posterior region of the notochord could be obtained. (A) A schematic drawing of an embryo at 19 hpf. (B) A fluorescence image of the posterior region (indicated in A) of the notochord of a Cecyil embryo at 19 hpf. (C) A fluorescence image of the posterior region (indicated in A) of the notochord of a Cecyil2 embryo at 19 hpf. (D) A schematic drawing of an embryo at 20 hpf. (E) A fluorescence image of the posterior region (indicated in D) of the notochord of a Cecyil embryo at 20 hpf. (F) A fluorescence image of the posterior region (indicated in D) of the notochord of a Cecyil2 embryo at 20 hpf. (G) A schematic drawing of an embryo at 22 hpf. (H) A fluorescence image of the posterior region (indicated in G) of the notochord of a Cecyil embryo at 22 hpf. (I) A fluorescence image of the posterior region (indicated in G) of the notochord of a Cecyil2 embryo at 22 hpf. (J–M) A Cecyil embryo at 18 hpf was treated with 400 μM EdU for 1 h and then fixed with 4% PFA. Alexa647-azide was used to visualize EdU incorporation. Fluorescence images of the notochord of a Cecyil embryo at 18 hpf for G1 marker (red) (J), S/G2/M marker (green) (K), incorporated EdU (white) (L), and their merge (M). [Scale bar, 50 μm (A–I); 100 μm (J–M.] |
|
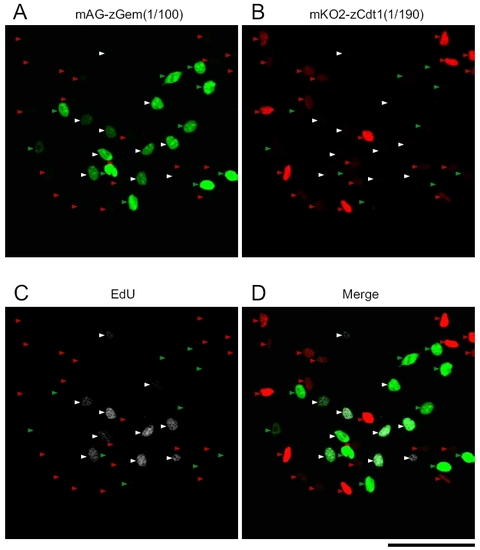
Characterization of zFucci for cell cycle progression. (A–D) A wide field image of Cecyil cells expressing zFucci: mKO2-zCdt1(1/190) and mAG-zGem(1/ 100) and detection of incorporated EdU. Red, green, and white arrows indicate G1, S, and G2 phases, respectively. (Scale bar, 100 μm.) |
|
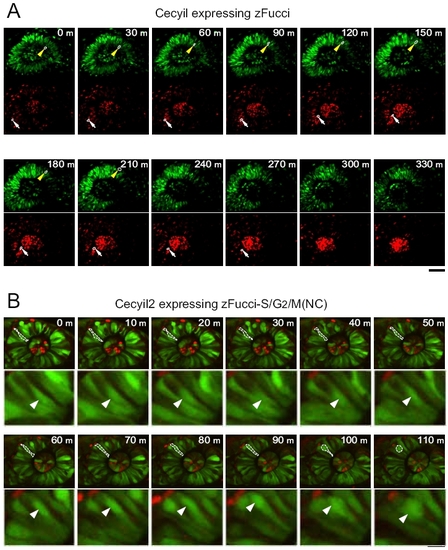
Interkinetic nuclear migration in the retina at early stages. An embryo at 22 hpf was anesthetized with 0.00168% Tricaine and embedded in 0.3% agar on a culture dish with the eye facing up. Fluorescence images were collected using an Olympus FV1000 upright confocal microscope to visualize interkinetic nuclear migration, namely, the apical-basal movement of nuclei in phase with the cell cycle. The developing lens at the center of the eye showed intense orange signal, suggesting early differentiation of the lens. (A) Fluorescence images of an eye from a Cecyil embryo expressing zFucci: mAG-zGem(1/100) (Top) and mKO2-zCdt1(1/190) (Bottom). Each nucleus in the sheet emitted either green or orange fluorescence. (Scale bar, 50 μm.) (B) Fluorescence images of an eye from a Cecyil2 embryo expressing zFucci-S/G2/M(NC): mAG-hGem(1/60) and mKO2-zCdt1(1/190) (merged). (Scale bar, 50 μm.) |