- Title
-
DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish
- Authors
- Takeda, Y., Mishima, Y., Fujiwara, T., Sakamoto, H., and Inoue, K.
- Source
- Full text @ PLoS One
|
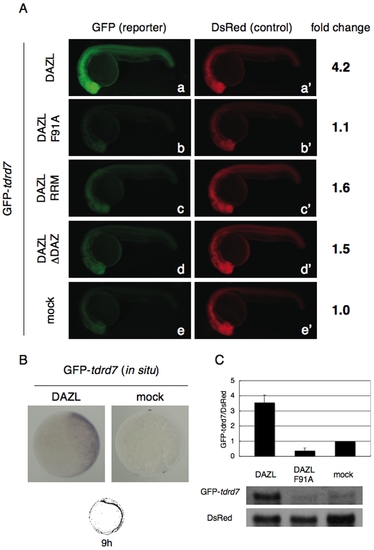
Activation of GFP-tdrd7 expression by DAZL. (A) GFP-tdrd7 and DsRed mRNAs were co-injected with the mRNAs encoding Myc-tagged DAZL, DAZL F91A, DAZL RRM, or DAZL ΔDAZ mRNA, or without any dazl mRNA (mock) at the one-cell stage. GFP and DsRed expression were analyzed at 24 hr post-fertilization (hpf). Fold change of normalized GFP fluorescence relative to mock control is shown on the right. (B) In situ hybridization of the injected embryos at 80% epiboly with an antisense probe for GFP. GFP-tdrd7 and DsRed mRNAs were injected with or without the mRNA encoding Myc-DAZL. (C) Northern blotting of the injected GFP-tdrd7 and DsRed mRNAs. The ratio of GFP mRNA to DsRed mRNA from three independent experiments is shown. |
|
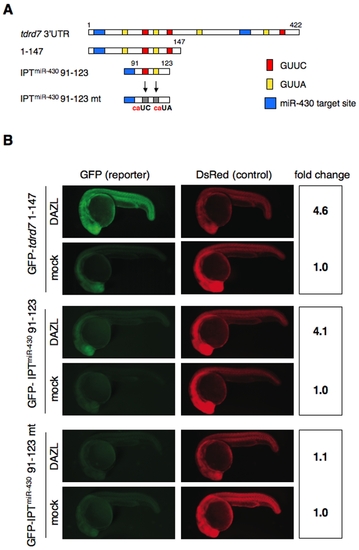
Identification of the cis-element required for the regulation by DAZL. (A) Schematic representation of tdrd7 3′UTR mutant constructs. Putative DAZL-binding sites (GUUC; red, GUUA; yellow), miR-430 target sites (blue), and mutated DAZL-binding sites (gray) are indicated. Nucleotide positions relative to the stop codon are shown above. The exogenous miR-430 target site (IPTmiR-430) in the IPTmiR-430 91–123 and IPTmiR-430 91–123 mt constructs were derived from the nanos1 3′UTR [5]. (B) Injection experiments of mutant GFP-tdrd7 mRNA and DsRed mRNA with or without the mRNA encoding Myc-DAZL. GFP and DsRed were analyzed at 24 hpf. Fold change of normalized GFP fluorescence relative to mock control is shown on the right. |
|
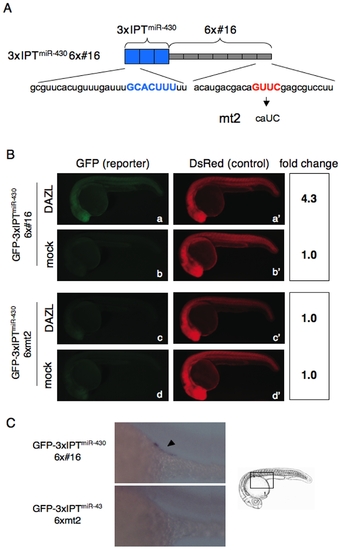
DAZL relieves the miR-430-mediated repression via binding to the cis-element GUUC. (A) Schematic representation of the 3′UTR of GFP-3xIPTmiR-430 6x#16 and GFP-3xIPTmiR-430 6xmt2. Sequences of IPTmiR-430 and #16 are shown below. The sequence that basepairs with miR-430 seed (blue) and the DAZL-binding sequence (red) are indicated. (B) Injection experiments of GFP-3xIPTmiR-430 6x#16 (a and b) or GFP-3xIPTmiR-430 6xmt2 (c and d) with or without the mRNA encoding Myc-DAZL. DsRed mRNA was co-injected. GFP and DsRed expression were analyzed at 24 hpf. Fold change of normalized GFP fluorescence relative to mock control is shown on the right. (C) In situ hybridization of the embryos injected with GFP-3xIPTmiR-430 6x#16 or GFP-3xIPTmiR-430 6xmt2 with a GFP probe at 24 hpf. The boxed region in the schematic representation was enlarged. Arrowhead indicates the PGCs. |
|
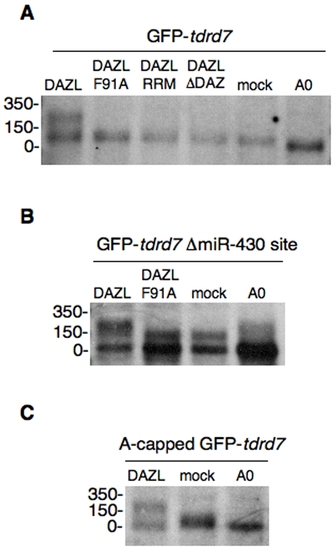
DAZL induces mRNA polyadenylation. Poly(A) length of GFP-tdrd7 (A), GFP-tdrd7 ΔmiR-430 site (B), or A-capped GFP-tdrd7 (C) in the presence or absence of DAZL proteins was analyzed by RNase H-poly(A) assay at 6 hpf. (A0) shows completely deadenylated fragments by RNase H digestion with oligo (dT). The position of the RNA size marker is shown on the left. |
|
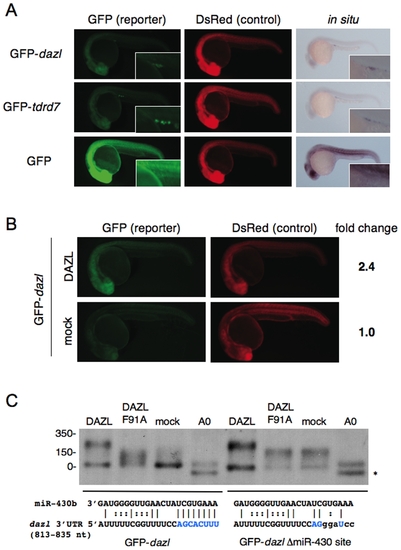
Regulation of dazl mRNA by miR-430 and DAZL. (A) GFP-dazl, GFP-tdrd7 or GFP mRNA was injected with DsRed mRNA. GFP and DsRed were analyzed at 24 hpf. Insets show the gonad region. The reporter mRNA was detected by in situ hybridization with a GFP probe. (B) GFP-dazl and DsRed mRNAs were injected with or without the mRNA encoding Myc-DAZL. GFP and DsRed were analyzed at 24 hpf. Fold change of normalized GFP fluorescence relative to mock control is shown on the right. (C) Poly(A) length of GFP-dazl and GFP-dazl ΔmiR-430 site mRNAs in the presence or absence of DAZL was analyzed by RNase H-poly(A) assay at 6 hpf. (A0) shows completely deadenylated fragments by RNase H digestion with oligo (dT). The position of the RNA size marker is shown on the left. Sequences of wildtype and mutated miR-430 target sites are shown below. Nucleotides that are complementary with miR-430 seed are shown in blue. Asterisk indicates a non-specific cleavage product of the dazl 3′UTR generated with oligo (dT). |
|
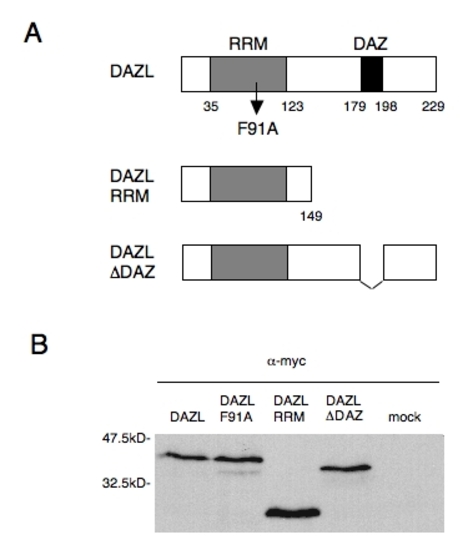
DAZL constructs used in this study. (A) Schematic representation of intact and mutant DAZL proteins. (B) Western blotting of Myc-tagged DAZL proteins expressed in the embryos with anti-Myc antibody. Molecular size markers are shown on the left. |
|
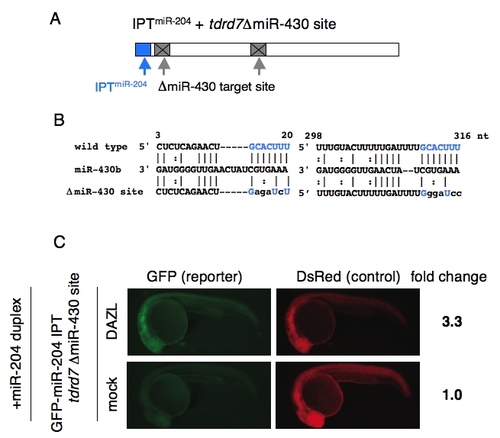
DAZL counteracts the miRNA repression. (A) Schematic representation of the 3′UTR of GFP-IPTmiR-204 tdrd7 ΔmiR-430 site mRNA. The target site of miR-204 (blue) and the mutated site for miR-430 (gray) are shown. (B) Sequences of wildtype and mutated miR-430 target sites in the tdrd7 3′UTR. Nucleotide positions relative to the stop codon were shown above. Nucleotides that basepair with miR-430 seed are indicated in blue. (C) The GFP-IPTmiR-204 tdrd7 ΔmiR-430 site and DsRed mRNAs were injected with or without the mRNA encoding Myc-DAZL at the one-cell stage. Subsequently, the miR-204 duplex was injected at the two-cell stage. GFP and DsRed were analyzed at 24 hpf. Fold change of normalized GFP fluorescence relative to mock control is shown on the right. |
|
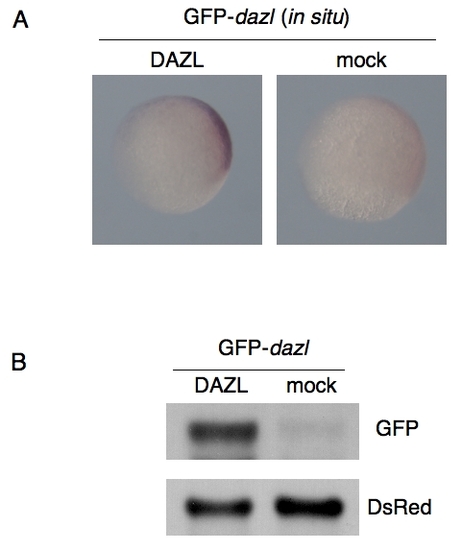
The effect of DAZL on GFP-dazl mRNA stability. (A) In situ hybridization of the injected embryos at 80% epiboly with an antisense probe for GFP. GFP-dazl and DsRed mRNAs were injected with or without the mRNA encoding Myc-DAZL. (B) Northern blotting of GFP-dazl and DsRed mRNAs injected with or without the mRNA encoding Myc-DAZL. |