- Title
-
Phoenix is required for mechanosensory hair cell regeneration in the zebrafish lateral line
- Authors
- Behra, M., Bradsher, J., Sougrat, R., Gallardo, V., Allende, M.L., and Burgess, S.M.
- Source
- Full text @ PLoS Genet.
|
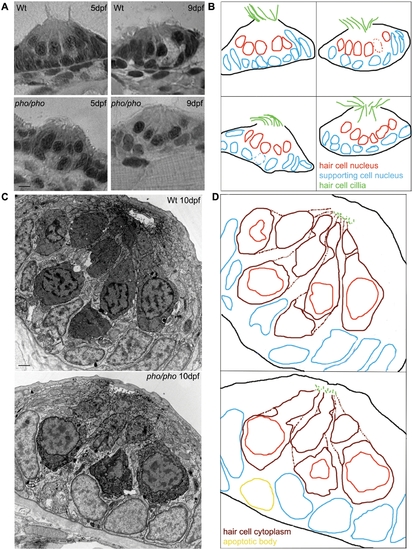
The development of the neuromasts in the lateral line is normal in the phoenix mutant larvae. (A) Semi-thin sections showing wild-type (top panels) and mutant neuromasts (bottom panels) in 5dpf (left panels) and 9dpf (right panels) larvae. (B) Camera lucida drawing for each section, highlighting the hair cells nuclei (red) and their cilia (green) and the supporting cells nuclei (blue). (C) Ultra-thin sections viewed by Electron Microscopy (EM) of a wild-type (top panel) and a mutant (lower panel) neuromast in 10dpf larvae. The hair cells stain darker than the supporting cells. (D) Camera lucida drawings of the EM sections, highlighting the hair cell nuclei (red), cell bodies (dark red), and cilia (green). The nuclei of the supporting cells are highlighted (blue). One apoptotic body was visible in (yellow). - 5 microns in (A) and 1 micron in (C). |
|
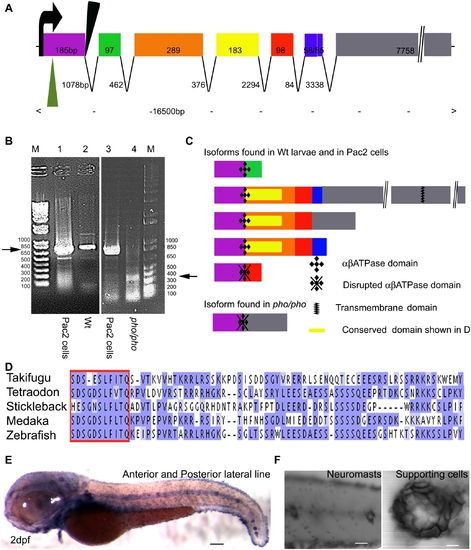
Phoenix is a gene without obvious homologs expressed in the supporting cells of the lateral line. (A) The coding region of the phoenix gene spans approximately 16.5kb with seven spliced exons. Exon 7 is an unusually long ORF of 7758bp. The retroviral integration in the zm allele (green triangle) is in the beginning of the first exon, probably leading to a null allele. In the hi43 allele, the retroviral integration (black triangle) is adjacent to the first splice donor site inducing an aberrant spliced mRNA (see [B] and [C]). The size of exons and introns are indicated. (B) RT-PCR on total RNA extracts of the zebrafish cell line PAC2 (lanes 1 and 3) and 7dpf wild-type (lane 2) and hi43 mutant allele (lane 4) larvae. The expected band of 835bp (left ->) was detected in the PAC2 cell line (lane 1 and 3) and in the wild-type larvae (lane 2). However, in the hi43 mutant lane there is a new 368 bp band representing an aberrant splice event from exon 1 to exon 7 (right ->). (C) We identified five splicing variants from wild-type larvae and the PAC2 cell line and one in the hi43 mutant allele larvae. In the mutant mRNA, the ab ATPase domain (plusses) is disrupted by the aberrant splice (diamonds). A putative transmembrane domain is found in exon 7 (zig-zag bar). The location of the conserved domain described in (D) is shown (yellow rectangle). (D) Alignment that shows the region of homology for putative phoenix genes in four other fish species. One particular stretch seems to be strongly conserved in all fish species, the short SDS- X(3)-SLF-[ILV]-TQ sequence (red box). The remaining sequence for all predicted proteins shows little to no significant identity, beyond enrichment for proline and lysine residues. (E) In situ hybridization with an antisense probe directed against the phoenix gene. At 2dpf the expression was found in all the neuromasts of the anterior and posterior lateral line. (F) Higher magnification of two trunk neuromasts (left panel) and a single neuromast (right panel), showing the typical ring-like staining restricted to the supporting cells. - 100 microns in E, 50 microns in (F, left panel) and 10 microns in (F, right panel). |
|
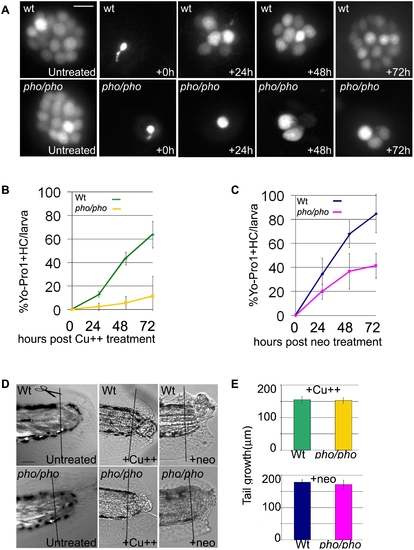
Lateral line hair cell regeneration is severely reduced in phoenix mutants but regeneration in other tissues is not affected. (A) Neuromasts of the lateral line are stained with YoPro 1, which exclusively stains hair cells. We treated wild-type (top panels) and pho/pho larvae (bottom panels) at 5dpf with copper (10mM). We monitored regeneration of hair cells over the following three days, at +24, +48, and +72h. As a reference, 5dpf wild-type and mutant neuromasts are shown just prior to treatment and just after the treatment (untreated and +0h, respectively, in the first and second columns from the left). (B) and (C) The number of hair cells was counted in untreated wild-type and mutant larvae at 5dpf (n = 39/53), 6dpf (n = 37/37), 7dpf (n = 23/22), and 8 dpf (n = 20/15). In each larva, we counted ten head neuromasts and all the tail neuromasts. We counted hair cells in healthy surviving larvae after 10 mM copper (B) or 200 mM neomycin (C) treatments. In (B), we counted wild-type (green line) and mutant (yellow line) larvae at +24h, (n = 40/36), +48h (n = 23/32), and +72h (n = 25/19), respectively, after copper treatment. In (C), we counted wild-type (blue line) and mutant (pink line) larvae at +24h, (n = 40/36), +48h (n = 23/32), and +72h (n = 25/19), respectively, after neomycin treatment. The numbers are presented as a percentage of regenerated hair cells in treated larvae, 100% being the respective number of hair cells at equivalent untreated wild-type and mutant stages. (D) We monitored the regeneration of tails in wild-type (top panels) and mutant (bottom panels) larvae. In untreated larvae (left panels), we amputated the tail at 3dpf and monitored the regeneration until 9dpf in wild-type (n = 8) and mutant (n = 20). We monitored the partial regeneration of tails amputated at 5dpf after copper (middle panels) and neomycin treatment (right panels). (E) Quantification of the tail regeneration after amputation at 5dpf after the copper treatment (upper graph), as measured in 8dpf wild-type (green, n = 11) and pho/pho (yellow, n = 10) larvae and after neomycin treatment (lower graph), as measured in 9dpf wild-type (dark blue, n = 6) and mutant (pink, n = 7) larvae. – 10 microns in (A) and 50 microns in (D). |
|
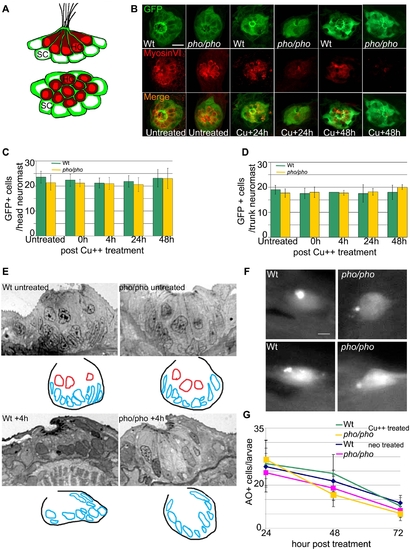
Supporting cells are present and do not die at a higher rate because of the treatments in the phoenix mutant neuromasts. (A) Schematic drawings of a side view (top panel) and a dorsal view (bottom panel) of a neuromast, illustrating the distribution of the two cell types. The hair cells (red) are centrally located and engulfed in a thin cytoplasmic furrow projected apically by the underlying supporting cells (green). (B) Immuno-labeled neuromasts with an antibody against GFP (top panels) staining the supporting cells and an antibody against Myosin VI (middle panels) staining the hair cells. Merged images are presented in the bottom panels. The SCM1 transgenic line, which we crossed into the phoenix mutant background, is expressing GFP exclusively in the supporting cells of the lateral line. We imaged transgenic wild-type (from the left, columns 1, 3, and 5) and transgenic phoenix mutant (columns 2, 4, and 6) larvae. Columns 1 and 2 are untreated 5dpf larvae. Columns 3 and 4 are larvae 24h post copper treatment. Columns 5 and 6 are larvae 48h post copper treatment. (C) Quantification of the number of GFP positive supporting cells respectively, in untreated (n = 18/21) and in +0h (n = 12/9), +4h (n = 15/17), +24h (n = 15/12), and +48h (n = 15/18) post copper treated transgenic wild-type (green bars) and transgenic phoenix mutant (yellow bars) head neuromasts. (D) Trunk neuromasts in wild-type and phoenix mutant transgenic larvae have been counted at the same stages, respectively untreated (n = 3/4), +0h (n = 4/3), +4h (n = 1/4), +24h (n = 2/5), and +48h (n = 2/3). (E) Semi-thin sections through wild-type (left panels) and mutant (right panels) neuromasts in untreated (top panels) and + 4h post neomycin treated (lower panel) 5dpf larvae. We used between 13 and 20 successive sections to reconstruct each wild-type and mutant neuromast (n = 1/2 untreated, n = 6/6 at +4h, respectively). We show one representative wild-type and mutant section at each stage. Under each section, a camera lucida sketch is showing the limits of the neuromast and the outlines of the nuclei (red for hair cells, blue for supporting cells). (F) Live images of two wild-type (left panels) and two mutant (right panels) neuromasts after AO stainings in larvae, 24h post copper treatment. (G) Scoring of the number of acridine orange (AO) positive cells in the lateral line/larvae, 24, 48, and 72h post copper (Cu++) or neomycin (neo) treatments. For the copper treatment, we monitored wild-type (green line) and mutant (yellow line, n = 22/13 at +24h, n = 14/12 at +48h, and n = 18/6 at +72h, respectively) larvae. For the neomycin treatment, we monitored wild-type (blue line) and mutant (pink line, n = 12/10 at +24h, n = 11/14 at +48h, and n = 10/11 at +72h, respectively) larvae. – 10 microns in (B) and (F) and 5 microns in (E). |
|
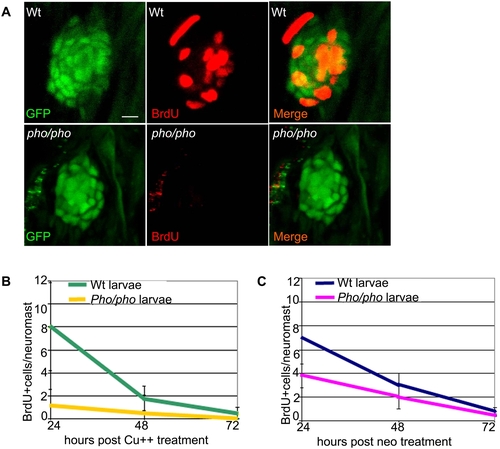
Supporting cells do not divide as efficiently after hair cell damage in the phoenix mutant neuromast. (A) BrdU staining (red, in the middle and right panels) in the SCM1::GFP transgenic wild-type (top) and the SCM1::GFP transgenic phoenix mutant neuromast (bottom) of recovering larvae (n = 20/20), 24h after copper treatment. The supporting cells are counterstained with GFP antibodies (green in left and right panels). (B) Quantification of the BrdU positive cells/neuromast in wild-type (green line) and mutant (yellow line) recovering neuromasts, at +24h (n = 11/10), +48h (n = 10/6), and +72h (n = 7/6) after copper treatment. Larvae are exposed to BrdU for 6h prior to being sacrificed. (C) Quantification of the BrdU positive cells/neuromast in wild-type (blue line) and mutant (pink line) recovering larvae, at +24h (n = 22/40), +48h (n = 20/33), and +72h (n = 13/32) after neomycin exposure. – 10 microns in (A). |
|
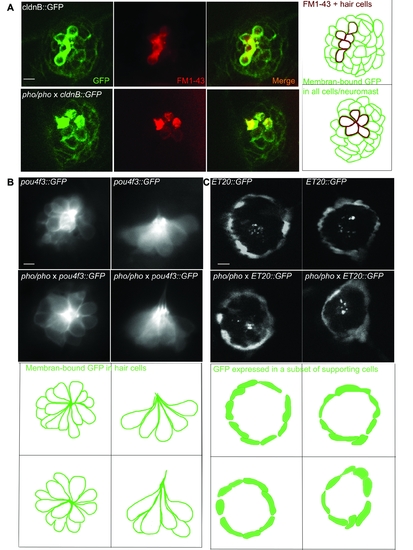
Live imaging of transgenic lines crossed into the phoenix background show no differences from wild-type larvae. A. We crossed the phoenix mutant line into the cldnB::GFP transgenic background, which expressed GFP (green in first and third panels) in all cells of the neuromast. We stained untreated 5 dpf larvae with FM1-43 (red in second and third a panel). Shown are images of live wild-type (top panels) and phoenix (bottom panels) neuromasts. A camera lucida drawing of each merged picture is highlighting the GFP positive cells contours (green) and the FM1-43 positive cells (red) in the fourth panel. B. We crossed the phoenix mutant line into the pou4f3::GFP transgenic background, which expressed GFP in all of the neuromast's hair cells. Dorsal views (left panels) and lateral views (right panels) of wild-type (top panels) and pho/pho (bottom panels) are shown. A camera lucida drawing of each panel is outlining the GFP positive hair cells of each respective 41 panel. C. We crossed the phoenix mutant into the ET20::GFP transgenic background, which expressed GFP in a subset of supporting cells known as the mantle cells. Two representative examples of wild-type (top panels) and of phoenix mutant (bottom panels) neuromasts are shown. A camera lucida drawing of each panel is outlining the GFP + cells of each respective panel. - 10 microns in all panels. |
|
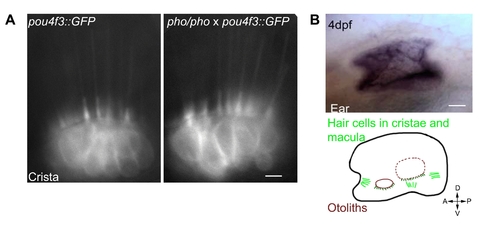
Hair cells seem unaffected in the phoenix mutant ear. Wild-type expression of the phoenix mRNA and protein. A. Live images in pou4f3;;GFP transgenic animals of hair cells in a crista of a wild-type (left panel) and a phoenix mutant (right panel) ear. B. In situ hybridization with an antisense probe against phoenix in the inner ear of a 4dpf old embryo (top panel). A camera lucida drawing (lower panel) shows the positions of the two otoliths (red), of the cristae's (light green) and the maculae's (dark green) hair cells. The AP/DV ventral orientation is indicated. C. Immunofluorescence on cultured zebrafish cells (Pac2) using EX1 (green in the top and bottom left panels) or EX7 (green in the top and bottom right panels) antibodies. To highlight the ER, we co-stained with an antibody against PDI (ER-associated protein disulfide isomerase, red in middle and bottom left and right panels). DAPI was used to counterstain nuclei (blue in the two bottom left and right panels). Merged images (bottom, left and right panels) showed a cytoplasmic staining of phoenix, which was more concentrated in the perinuclear region. It appeared excluded from the ER, as there was no co-localization with DPI. Note how phoenix protein was upregulated in cells preparing to enter division (white arrow in the top right panel). D. Western blot on PAC2 cell extracts with two rabbit polyclonal antibodies, raised against an epitope encoded by exon 1 (EX1) or by exon 7 (EX7). A common band in the range of 95KDa was found with both antibodies (black arrow). - 10 microns in A, 50 microns in B and 3 microns in C. |