- Title
-
Inca: a novel p21-activated kinase-associated protein required for cranial neural crest development
- Authors
- Luo, T., Xu, Y., Hoffman, T.L., Zhang, T., Schilling, T., and Sargent, T.D.
- Source
- Full text @ Development
|
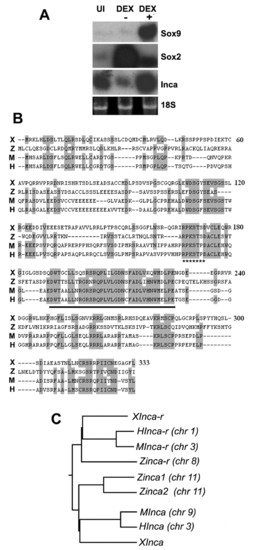
Inca is a novel Tfap2a target in the NC. (A) Animal caps were injected with Wnt3a, Chd and GR-AP2a mRNA, treated with 10 μM dexamethasone (DEX+) or solvent alone (DEX-), and subjected to northern blot analysis using probes as indicated, with 18S rRNA as a loading control. UI, uninjected. (B) Predicted protein sequence alignment for Xenopus (X), zebrafish (Z), mouse (M) and human (H) Incas. Identical residues are shaded. The conserved 38-residue inca-box is underlined, and the 14-3-3 binding site is indicated with star underline. (C) Dendrogram generated using ClustalW (http://clustalw.genome.jp). Accession Numbers are: Xenopus IncaA, BJ092564. Mouse, AK076092. Human, NM_203370. Zebrafish inca1, CK018555; inca2, BM071087. Human Inca-r, BC031558. Mouse Inca-r, NM_175398. Zebrafish inca-r, BI885065. Xenopus (tropicalis) Inca-r, CR761080. chr, chromosome number. |
|
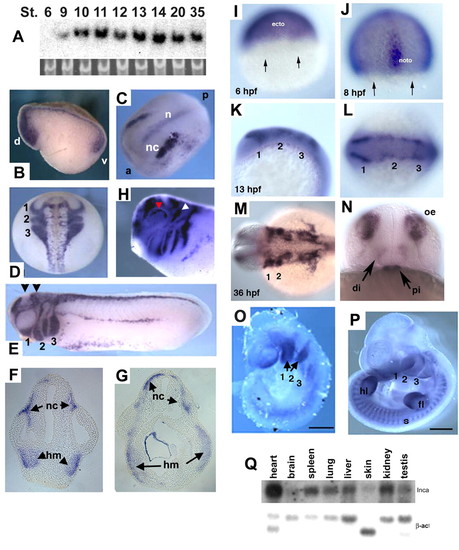
Expression patterns in Xenopus, zebrafish and mouse embryos. (A) Developmental northern blot of Xenopus Inca, with 18S RNA as a loading control. Nieuwkoop-Faber stages are indicated. (B-H) Whole-mount in situ hybridization for Inca in Xenopus embryos at stages 10.5-32. (B) Stage 10.5, sagittal section. d, dorsal. v, ventral. (C) Stage 14, dorsal view of expression in cranial NC (nc) and notochord (n). a, anterior. p, posterior. (D) Stage 19, dorsal view of expression in cranial NC migration streams, labeled as 1-3. (E) Stage 25, lateral view; arrowheads indicate approximate section levels in F and G. NC migration streams into pharyngeal arches 1-3 as indicated. (F,G) Transverse sections of embryo in E show Inca expression in head mesenchyme (hm) and NC (nc). (H) Stage 32, lateral view of the head. White and red arrowheads indicate expression in trigeminal ganglion and eye, respectively. (I-N) Whole-mount in situ hybridization for inca1 in zebrafish embryos. (I) 6 hpf, lateral view, dorsal to the right. Expression is restricted away from the margin (arrows), in future ectoderm (ecto). (J) 8 hpf, dorsal view. noto, notochord. (K,L) 13 hpf, lateral and dorsal views, respectively, of inca1 expression in premigratory NC. Numbers indicate presumptive pharyngeal arches 1-3. (M,N) 36 hpf, dorsal and face-on views, showing expression in the pharyngeal arches (1,2), diencephalon (di), pituitary (pi) and olfactory epithelia (oe). (O,P) Inca expression in mouse embryos at E9.5 (O) and E11.5 (P) in pharyngeal arches (numbered 1-3), somites (s) and fore-limb (fl) and hind-limb buds (hl). (Q) Northern analysis of adult mouse tissue RNAs probed with Inca and beta-actin as control. Scale bars: 500 μm in O; 1 mm in P. |
|
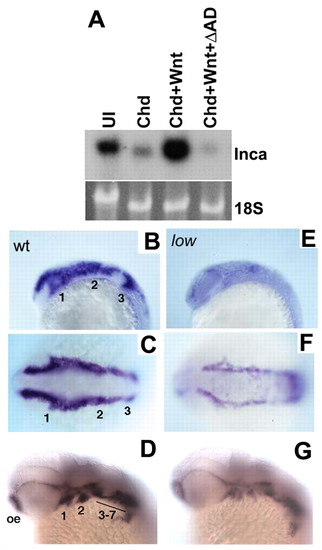
Inca expression in NC depends on Tfap2a. (A) Northern analysis of animal cap RNA from embryos injected with Chd, Chd+Wnt3a, or Chd+Wnt3a+dnTfap2a (Chd+Wnt+ΔAD) and probed for Xenopus Inca, with 18S rRNA as a loading control. Expression induced by Chd+Wnt is blocked by dnTfap2a. (B-G) Whole-mount in situ hybridizations showing zebrafish inca1 expression in wild type (B-D) and low mutants (E-G). B,E are lateral views and C,F dorsal views at the 10-somite stage. D,G are lateral views at 36 hpf. inca1 expression is reduced at 10 somites and 36 hpf. Numbers indicate presumptive pharyngeal arches. oe, olfactory epithelia. EXPRESSION / LABELING:
|
|
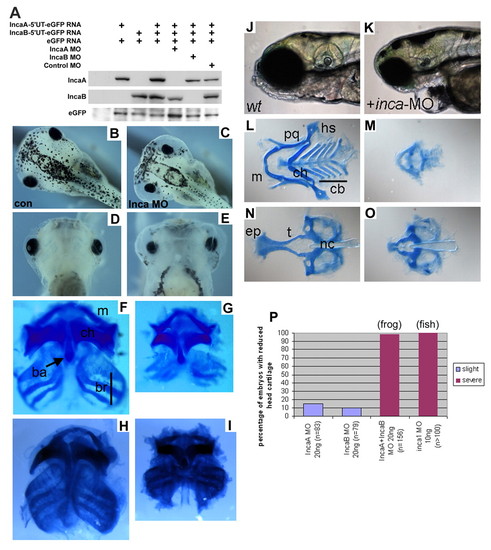
Inca is required to form the NC-derived head skeleton in both frog and fish. (A) Western blot analysis showing that Xenopus Inca MOs efficiently inhibit translation of injected Inca-GFP mRNA. (B-I) Stage 45 Xenopus tadpoles injected with control MO (B,D,F) or Inca MO (C,E,G) and shown in dorsal (B,C) and ventral (D-I) views either live (B-E) or stained with Alcian Blue for cartilage (F-I). Similar results were obtained with two independent, non-overlapping sets of Inca MOs (H, 20 ng control MO; I, 20 ng IncaA MO2+IncaB MO2). (J-O) 5-day-old control zebrafish larvae (J,L,N) or inca1 MO morphants (K,M,O) shown live in lateral view (J,K) or Alcian Blue-stained in ventral view (L-O). All cartilage elements are reduced by Inca knockdown in both species. ba, basihyal; br, branchial/gills; cb, ceratobranchial; ch, ceratohyal; ep, ethmoid plate; hs, hyosymplectic; m, Meckel's; nc, notochord; pq, palatoquadrate; t, trabeculae. (P) Frequency of head cartilage phenotype with Xenopus IncaA MO or IncaB MO alone or combined IncaA+IncaB, and Zebrafish inca1-MO injected embryos. PHENOTYPE:
|
|
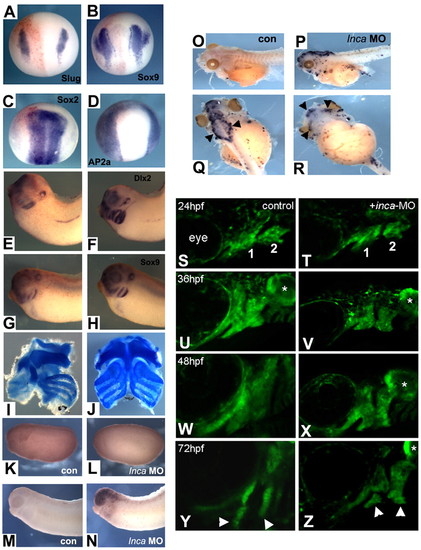
Inca is required for cranial NC specification and survival after migration into the arches in Xenopus and zebrafish. Embryos were co-injected into one cell at the two-cell stage with Inca MO, together with lacZ RNA as a lineage tracer and assayed for gene expression by whole-mount in situ hybridization at stage 14 (A-D, injected sides are on the left) and stage 32 (E-H; E,G, injected sides) with probes indicated in the panels. In no case was any change in gene expression observed on the injected side [(A) Slug, n=83; (B) Sox9, n=42; (C) Sox2, n=47; (D) AP2a, n=54; (E) Dlx2, n=63; (G) Sox9, n=78]. At tadpole stage, cranial cartilage was eliminated on the injected side (left side in I). Control MO injection had no effect (J). (K-R) TUNEL staining of embryos injected with control MO (K,M,O) or Inca MO (L,N,P-R), cultured to stage 21 (K,L), stage 30 (M,N) or stage 45 (O-R). Q and R are dorsal and ventral views, respectively, of the tadpole shown in P. Black arrowheads indicate strong TUNEL signal in neurocranium (Q) and jaw cartilage (R). (S-Z) Confocal images of sox10:egfp expression in cranial NC cells of controls (S,U,W,Y) and inca1 morphants (T,V,X,Z). Lateral views, anterior to the left. Stages are indicated in the panels. First and second pharyngeal arches are indicated in S,T, and derived cartilage elements by arrowheads in Y,Z. Asterisks indicate expression of sox10:eGFP in the otic vesicle. PHENOTYPE:
|
|
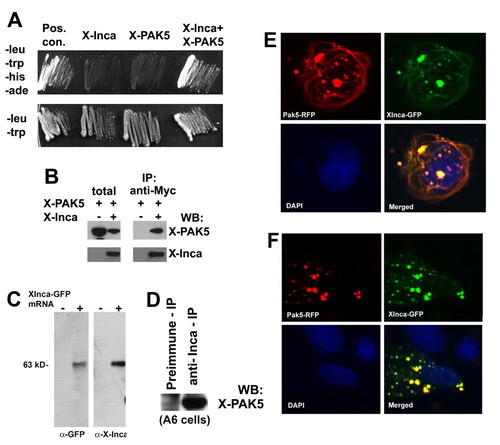
Inca interacts with PAK5. (A) Selective medium streaks of yeast cells expressing Xenopus Inca and PAK5, either alone or together, as indicated. Inca+PAK5 allows growth on medium lacking leucine, tryptophan, histidine and adenine. Positive controls include transfection with vectors containing T antigen and p53 (Clontech). Streaks on -leu/-trp (lower panel) shown as a control for transfection. (B) Extracts prepared from HEK293 cells transfected with expression plasmids encoding a PAK5-GFP fusion or a Myc epitope-tagged Inca separately or together, immunoprecipitated with anti-Myc, followed by western blot with antibody for GFP or Myc. Lanes labeled as total are from lysates prior to immunoprecipitation. PAK5-GFP precipitates with the anti-Myc antibody, but only when Myc-tagged Inca is present in the extract. (C) Anti-IncaA peptide antibody specificity. Fertilized eggs were injected with 1 ng of synthetic mRNA encoding XInca-GFP, and cultured to stage 20. Detergent (1% NP40)-solubilized protein was extracted with 1,1,2-trichlorotrifluoroethane (Freon) to remove yolk and the equivalent of two embryos analyzed by SDS-PAGE/western blot. Both anti-GFP and anti-Inca recognized a single band of the correct molecular weight (∼63 kDa). (D) Extract from untransfected Xenopus A6 cells immunoprecipitated with preimmune serum or anti-Inca, followed by western blot using antiserum raised against PAK5 (residues 122-224, a generous gift of N. Morin). Immunoprecipitation with anti-Inca enriches the PAK5 signal several-fold compared with preimmune serum, indicating Inca-PAK5 interaction. (E) Fluorescent images of a CHO cell cotransfected with plasmids encoding a PAK5-RFP fusion and Xenopus Inca-GFP fusion showing extensive overlap. (F) Fibrous structures positive for Inca and PAK5 are nocodazole-sensitive. CHO cells transiently transfected with PAK5-RFP and XInca-GFP were treated for 1 hour with 3 ng/ml nocodazole (Sigma), then fixed with methanol and photographed using an inverted fluorescence microscope. The fibers visible without nocodazole treatment (E) have disappeared. |
|
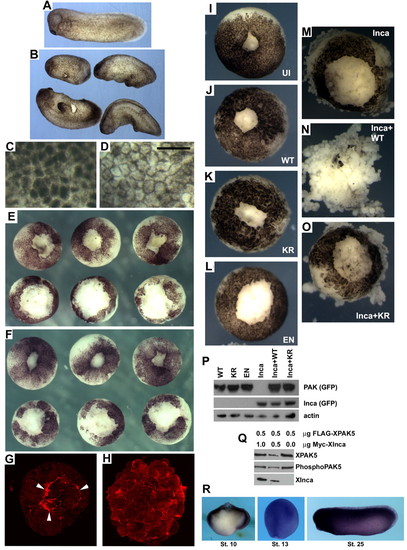
Inca misexpression disrupts gastrulation, pigmentation and wound healing in Xenopus. (A) Uninjected stage 25 control embryo. (B) Siblings injected with 250 pg IncaA mRNA have shorter body axes and numerous protuberances. (C,D) High-magnification views of ectoderm at stage 8 from uninjected (C) and IncaA mRNA-injected (250 pg; D) embryos showing relocation of cortical pigment to the cell periphery. Scale bar: 100 μm. (E,F) Vegetal explants after animal cap removal and culture for 5 (E) or 25 (F) minutes in 0.3xMMR. Upper row, uninjected; lower row injected with 250 pg IncaA mRNA, showing delayed healing. (G,H) Confocal z-stack images of animal caps from uninjected (G) and IncaA mRNA-injected (H) embryos after 5 minutes of healing in 0.3xMMR. White arrowheads in G indicate purse-string actin-filament structures missing in embryos overexpressing Inca. (I-O) Synergistic effect on wound healing of PAK5 and Inca. Embryos injected with mRNA encoding wild-type PAK5-GFP (J), kinase-dead PAK5-GFP (PAK5/KR; K), constitutively active PAK5-GFP (PAK5/EN; L), IncaA-GFP (M), a mixture of IncaA-GFP and PAK5-GFP (N) or a mixture of IncaA-GFP and PAK5/KR-GFP (O). All PAK5-GFP mRNAs injected at 1 ng, IncaA-GFP mRNA at 250 pg. Vegetal explants were allowed to heal for 25 minutes. Combining Inca+wild-type PAK5 greatly delays wound healing and leads to dissociation (N). PAK5/KR does not exhibit this synergistic effect (O). (I) Uninjected control embryo. Results from these experiments are summarized in Table 1. (P) PAK5-GFP mRNAs were equally translated, as judged by western blot using antibody to GFP. (Q) Co-expression with Inca does not increase PAK5 phosphorylation. Western blots of extracts made from HEK293 cells transfected with 0.5 µg FLAG-tagged XPAK5 plasmid and 0, 0.5 or 1.0 µg of Myc-tagged Xenopus Inca. The ratio of total XPAK5 signal (FLAG antibody) to the level of phosphorylation of regulatory serine residue 474 (phospho-PAK4) was not significantly different in any of the samples. (R) Xenopus PAK5 expression. Whole-mount in situ hybridization at stages 10, 13 and 25. Stage 10 embryos were bisected in the sagittal plane prior to hybridization. PAK5 expression is broad in ectoderm and mesoderm at early stages and essentially ubiquitous later in development. |