- Title
-
Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon
- Authors
- Davidson, A.E., Balciunas, D., Mohn, D., Shaffer, J., Hermanson, S., Sivasubbu, S., Cliff, M.P., Hackett, P.B., and Ekker, S.C.
- Source
- Full text @ Dev. Biol.
|
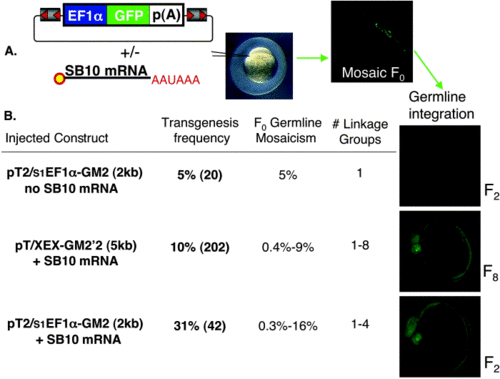
Injection scheme for the generation of SB transgenic lines. (A, left) Structure of a Sleeping Beauty transposon consisting of flanking IR/DR sequences (gray with orange triangles), a ubiquitous EF1α enhancer/promoter (blue) driving expression of a GFP reporter (green), and terminating with a SV40 poly(A) signal (white). In vitro-transcribed, capped SB10 transposase mRNA. The SB transposon is injected with or without SB10 mRNA into one-cell zebrafish embryos (A, middle), resulting in mosaic GFP expression at 24 hpf (A, far right). (B, left) Each injected transposon vector +/- SB10 mRNA is listed with the corresponding germline transmission and expression frequency, obtained as the percentage of F0 fish producing GFP-positive offspring. The number of fish screened is shown in parentheses. The F0 germline mosaicism rate is determined as the percentage of GFP-positive F1 offspring from a founder outcross. The number of linkage groups harboring a transposon insertion is also listed. For each experiment, embryos with germline integration of ubiquitous GFP expression at 25 hpf in the F2 (top and bottom) and F8 (middle) generations are shown under GFP fluorescence detection conditions. |
|
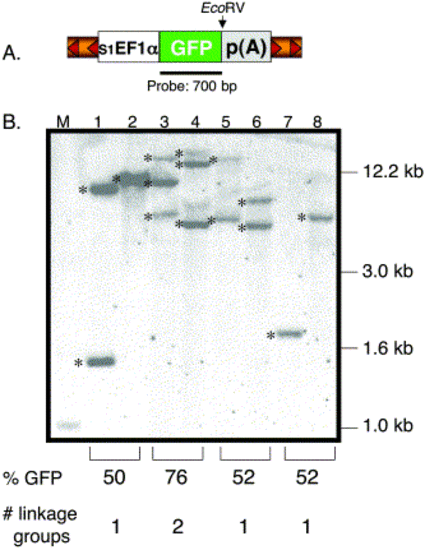
Southern blot analysis of transgenic zebrafish. (A) Structure of the pT2 transposon depicting the relative position of the GFP probe and EcoRV site. EcoRV cuts once in the pT2 transposon and NsiI is absent in the vector. (B). The EcoRV- or NsiI-digested genomic DNA was hybridized with the GFP-specific probe. Lanes 1, 3, 5, and 7 were digested with EcoRV and lanes 2, 4, 6, and 8 with NsiI. M, DNA marker lane. Shown are 6 independent insertions (*) from 3 individual F1 lines (lanes 3 and 4, lanes 5 and 6, lanes 7 and 8). The predicted number of linkage groups is based on the GFP expression data from the F1 outcrosses. One transgenic line previously shown to be carrying a single T-insertion was used as a control (lanes 1 and 2). |
|
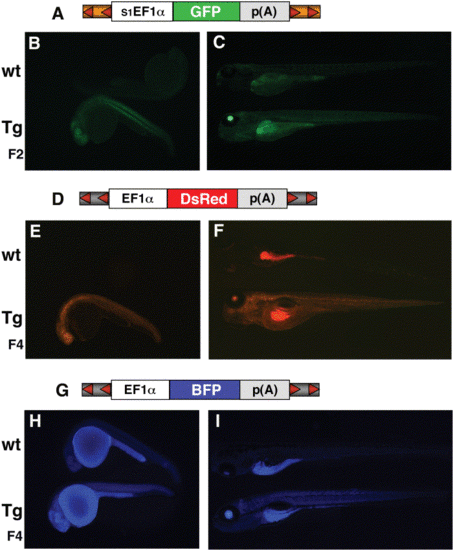
Ubiquitous expression of GFP, BFP and DsRed from transposon vectors. A schematic representation of each transposon used to generate the transgenic lines (Tg) is located above each set of fluorescent embryos in (A), (D), and (G). The T2-based vector expressing GFP is under the control of s1EF1α (A), and the T-based vectors expressing DsRed-1 (D) and BFP (G) are under the control of EF1α enhancer/promoter (XEX). Multiple generations expressing GFP (B, C), DsRed (E, F), and BFP (H, I) are shown at 25 hpf (B, E, H) and 5 dpf (C, F, I) along with a wild-type (wt) embryo. |
|
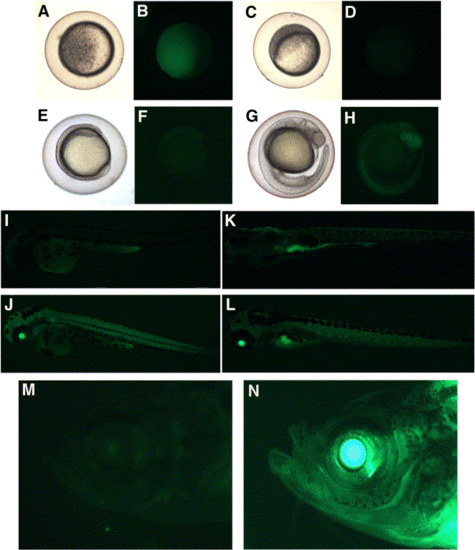
Expression analysis of T/XEX-GM2′2. Progeny from a homozygous transgenic female outcross (A, B) and a homozygous transgenic male outcross (C, D, E–H, J, L) are compared with wt embryos (I, K). Embryos were analyzed at the late sphere stage (A–D); 3- to 4-somite (E, F); 24 hpf (G, H); 48 hpf (I, J); 5 dpf (K, L), and homozygous adults (N) were compared with wt adults (M) for GFP expression. Images were captured under brightfield (A, C, E, G) and a bandpass GFP filter set (B, D, F, H, I, J, K, L, M, N). |
|
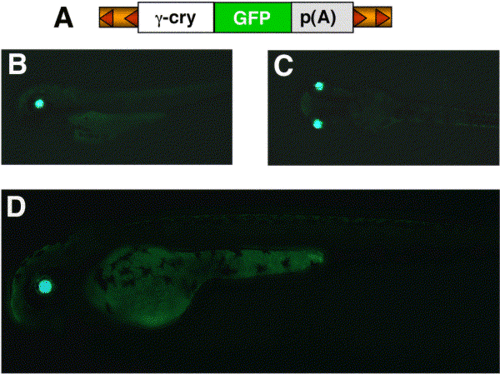
Tissue-specific expression from SB transposons. Coinjection of pT2 transposons (A) containing GFP under the control of an abbreviated X. laevis γ-crystallin promoter (see methods) and SB10 mRNA produces embryos with eye-specific GFP expression in F0 embryos at 48 hpf. (B) Lateral view. (C) Dorsal view. An F1 embryo expressing GFP specifically in the eye at 48 hpf is shown in (D). |
Reprinted from Developmental Biology, 263(2), Davidson, A.E., Balciunas, D., Mohn, D., Shaffer, J., Hermanson, S., Sivasubbu, S., Cliff, M.P., Hackett, P.B., and Ekker, S.C., Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon, 191-202, Copyright (2003) with permission from Elsevier. Full text @ Dev. Biol.