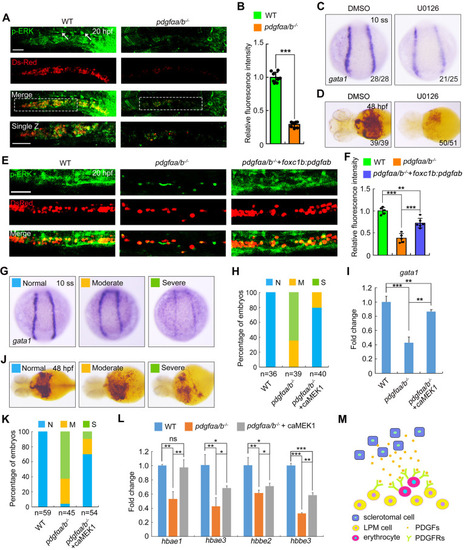
ERK1/2 functions downstream of PDGF signaling to promote primitive erythrocyte development. (A,B) Immunostaining of phosphorylated ERK1/2 (p-ERK) in wild-type (WT) and pdgfαa−/−;pdgfαb−/− (pdgfαa/b−/−) Tg(gata1:dsRed) embryos. (A) Representative confocal images. Single sections of the boxed areas are shown as enlarged images in the bottom panels. Lateral view with anterior to the left. The white arrows indicate endothelial cells of the DA. Scale bars: 50 μm. This experiment was repeated independently three times with similar results. (B) Average fluorescence intensity of p-ERK in the ICM region was calculated using ImageJ software, and the fluorescence intensity values relative to WT are shown as mean±s.d. (n=8). Two-tailed, unpaired Student's t-test was used to analyze the difference between groups. ***P<0.001. (C) In situ hybridization expression analysis of gata1 in WT embryos treated with 25 μM U0126 or DMSO as a vehicle control from the shield stage to the 10-somite stage (10 ss). Dorsal view. (D) Determination of hemoglobin levels by o-dianisidine staining in U0126-treated WT embryos at 48 hpf. Ventral view. Images shown in C and D are representative of the observed phenotypes. The number of embryos displaying each phenotype out of the total number assayed is indicated. (E,F) Immunostaining staining of p-ERK at 20 hpf in Tg(gata1:dsRed) background embryos. pdgfαa/b−/− embryos were injected with 30 pg foxc1b-pdgfαb-P2A-EosFP plasmids and 200 pg Tol2 transposase mRNA at the one-cell stage. WT and pdgfaαa/b−/− control groups were uninjected. (E) Representative confocal images. Lateral views. Scale bar: 50 μm. (F) The average fluorescence intensity of p-ERK in the ICM region was calculated using ImageJ software, and the fluorescence intensity values relative to WT are shown as mean±s.d. (n≥5) from three independent experiments. One-way ANOVA with Scheffe post-hoc test was used to analyze the statistical differences between groups. **P<0.01, ***P<0.001. (G-I) pdgfαa−/−;pdgfαb−/− embryos were injected with 20 pg caMEK1 mRNA at the one-cell stage and then harvested alongside uninjected WT and pdgfαa/b−/− control embryos for detection of gata1 expression by in situ hybridization at 10 ss. (G) Representative images of observed phenotypes. Dorsal view. (H) The percentage of embryos scored as being normal (N) or as having moderate (M) or severe (S) defects for the indicated treatment groups. (I) Quantitative real-time PCR was performed to examine the expression of gata1 in the indicated embryos, and the values are expressed as mean±s.d. Fold change relative to WT from three independent experiments. One-way ANOVA with Tukey post-hoc test to analyze the statistical differences between groups. **P<0.01, ***P<0.001. (J-L) Hemoglobin levels in embyros as described in G-I were examined at 48 hpf using o-dianisidine staining. (J) Representative images of phenotypes observed. Ventral view. (K) The percentage of embryos scored as being normal (N) or as having moderate (M) or severe (S) defects for the indicated treatment groups. (L) Quantitative real-time PCR was performed to examine the expression of hbae1, hbae3, hbbe2 and hbbe3, and the values are expressed as mean±s.d. Fold change relative to WT from three independent experiments. One-way ANOVA with Tukey post-hoc test was used to analyze the statistical differences between groups. *P<0.05; **P<0.01; ***P<0.001; ns, not significant. (M) A proposed model illustrating that PDGF ligands derived from sclerotome interact with and activate their receptors in the nearby lateral plate mesoderm, thereby supporting erythroid progenitor differentiation. LPM, lateral plate mesoderm.
|