Fig. 1
- ID
- ZDB-FIG-231108-45
- Publication
- Pfefferli et al., 2023 - Parallels between oncogene-driven cardiac hyperplasia and heart regeneration in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
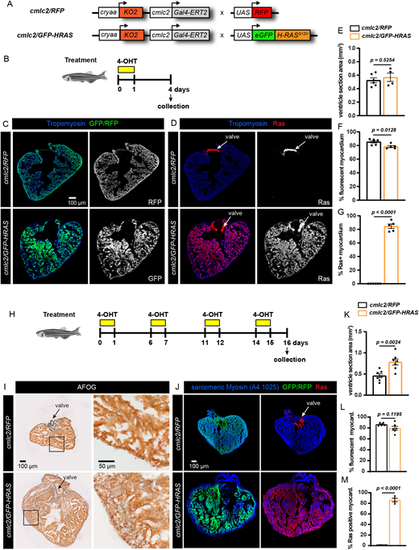
Inducible oncogenic HRASG12V leads to cardiac overgrowth in adult zebrafish. (A) Schematic representation of the transgenic strains used for inducible expression of oncogenic HRASG12V in the heart. The construct with cmlc2:Gal4-ERT2 is linked to a lens marker, cryaa:KO2, to facilitate identification of transgenic fish. These fish were crossed with either UAS:mRFP1 (cmlc2/RFP, control) or UAS:eGFP-HRASG12V (cmlc2/GFP-HRAS, an oncogenic protein fused with eGFP). (B) Experimental design with one treatment of 2.5 µM hydroxytamoxifen (4-OHT) for 18 h, followed by 3 days of recovery. (C,D) Transverse sections of ventricles isolated from fish in the experiment illustrated in B. GFP represents the endogenous fluorescence. RFP, RAS and Tropomyosin were detected by immunofluorescence. (E-G) Quantification of different parameters, as indicated on the y-axis, in the experiment presented in A-D. Each dot on the graph corresponds to one heart and it represents an average of three non-adjacent sections (spaced by ∼100 µm). Error bars indicate s.e.m. P-values are calculated using unpaired two-tailed t-test. n≥4. (H) Experimental design with four pulses of 2.5 µM 4-OHT treatment, each treatment was carried out for 18 h. (I,J) Transverse sections of ventricles isolated from fish in the experiment illustrated in H. (K-M) Quantifications of different parameters, as indicated on the y-axis, in the experiment presented in H-J. n≥4. Data are mean±s.e.m.; unpaired two-tailed t-test. |