|
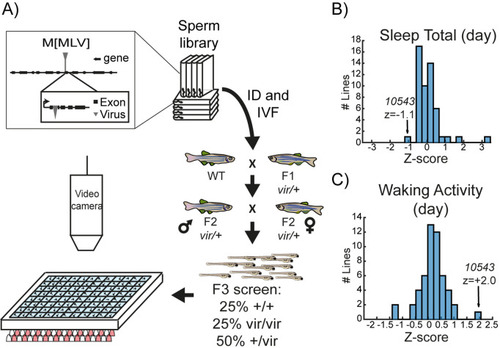
A viral insertion screen for sleep-wake regulators. (A) Schematic of screening strategy. Candidate genes were selected from a list of 904 mammalian genes encoding protein classes most often linked to behavioural regulation, including (1) genes previously implicated in sleep and circadian rhythms; (2) G-protein coupled receptors; (3) neuropeptide ligands; (4) channels; and (5) proteins involved in post-translational regulation, such as de-ubiquitinating enzymes (Figure 1—source data 1). tBLASTN of the human protein sequences identified 1162 zebrafish orthologs (Zv6), of which 702 (60.4%) had viral insertions mapped in the ‘Zenemark’ zebrafish viral insertion library (Varshney et al., 2013). Sperm harbouring viral insertions in 26 loci were successfully used for in vitro fertilisation and propagated to the F3 generation for screening. F3 larvae from single family F2 in-crosses were monitored on a 14 hr:10 hr light:dark cycle from 4 to 7 dpf using videography and genotyped at the end of the experiment. (B, C) Histogram of total daytime sleep (B) and average daytime waking activity (C) normalised as standard deviations from the mean (Z-score) of all the viral-insertion lines tested (including heterozygous vir/+ and homozygous vir/vir). Line 10543 (renamed dreammist) exhibited decreased daytime sleep and increased daytime waking activity.
|