Fig. 6
- ID
- ZDB-FIG-230516-25
- Publication
- Chen et al., 2022 - TGS1 impacts snRNA 3'-end processing, ameliorates survival motor neuron-dependent neurological phenotypes in vivo and prevents neurodegeneration
- Other Figures
- All Figure Page
- Back to All Figure Page
|
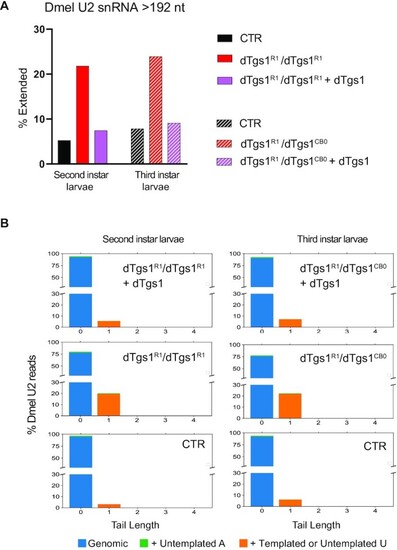
3′-Extended U2 snRNA molecules accumulate in Drosophila dTgs1 mutants. (A) Percentage of extended U2 snRNA molecules determined by 3′ RACE and sequencing on RNA from equally aged second or third instar mutant larvae, rescued mutant larvae (bearing a dTgs1 construct) and wild-type larvae (CTR). The most abundant population of U2 snRNAs is 192 nt long. Extended U2 snRNAs are longer than 192 nt and incorporate templated or untemplated nucleotides at their 3′ ends. Total reads > 2900. (B) Tail length and composition of the 3′ ends of U2 snRNAs for the second and third instar larvae described in (A). Numbers on the x-axis: additional nucleotides beyond the mature form of 192 nt (position 0). y-axis: percentage of total reads. |