Fig. 4
- ID
- ZDB-FIG-221214-63
- Publication
- Carrington et al., 2022 - A robust pipeline for efficient knock-in of point mutations and epitope tags in zebrafish using fluorescent PCR based screening
- Other Figures
- All Figure Page
- Back to All Figure Page
|
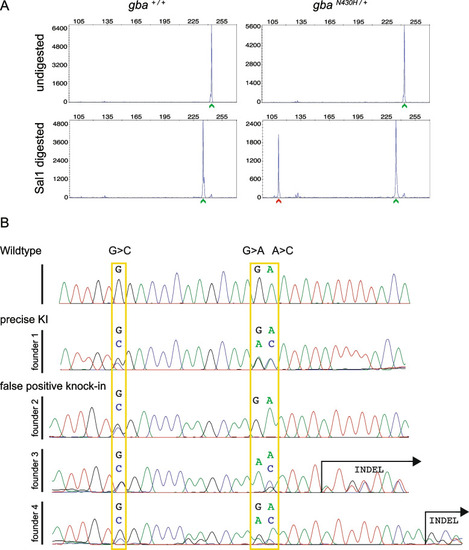
Germline screening data for knock-in of the point mutation in gba. A Representative plots of undigested and SalI digested fluorescent PCR products showing detection of the mutant allele (N430H) denoted by the red arrowhead in the progeny of a germline transmitting founder. In all plots, the X-axis represents the size of the peaks and the Y-axis shows the peak height. WT alleles (denoted by green arrowhead) are slightly smaller after digest (bottom panel) due to the restriction site in reverse primer used as internal control for successful restriction enzyme digestion. B Sequence chromatograms from representative embryos from each of the 4 founders positive for somatic knock-in compared with a WT embryo. Expected nucleotide modifications by knock-in of the ssODN are marked by yellow rectangles. Only founder 1 transmitted precise integration of the ssODN with all 3 modifications while the other 3 founders were false positives. Founder 2 had incomplete integration (the nucleotide for the restriction site was modified (G > C) but the desired (A > C) allele not changed) and founders 3 and 4 had indels |