- Title
-
A robust pipeline for efficient knock-in of point mutations and epitope tags in zebrafish using fluorescent PCR based screening
- Authors
- Carrington, B., Ramanagoudr-Bhojappa, R., Bresciani, E., Han, T.U., Sood, R.
- Source
- Full text @ BMC Genomics
|
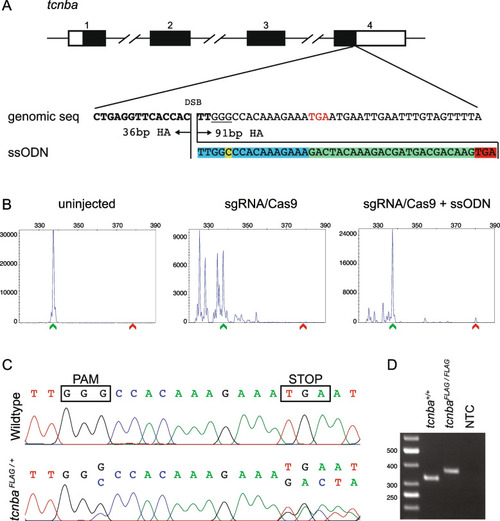
Design, screening, and validation of knock-in of FLAG tag at 3’ end of tcnba. A Schematic of tcnba genomic structure with coding region shaded in black (top panel), alignment of genomic sequence and ssODN template with direction of 36 bp and 91 bp homology arms (HA) marked. The ssODN contains a spacer (highlighted in blue) to maintain the ORF of tcnba as the double stranded break occurs 16 bp upstream of the stop codon (TGA, marked in red), a modification to the PAM site (G > C, highlighted in yellow) to prevent recutting after integration and the FLAG tag to be inserted (highlighted in green). B Representative CRISPR-STAT plots of uninjected, sgRNA/Cas9, and sgRNA/Cas9 plus ssODN injected embryos with X-axis showing the size of the peaks, Y-axis showing the peak height, and the size of the expected WT allele denoted by a green arrowhead. These plots were quantified for the presence of a peak generated by the insertion of ssODN (denoted by red arrowhead). C Sequence chromatograms showing WT and a positive F1 embryo to confirm germline transmission of knock-in sequence. D An agarose gel image showing expression of the FLAG tag at tcnba locus by RT-PCR. RNA from tcnba+/+ or tcnbaFLAG/FLAG embryos were used as template and water was used as the no template control (NTC). The original gel image is provided in additional file 2: Fig. 1 – Supporting data and its cropped version is shown here |
|
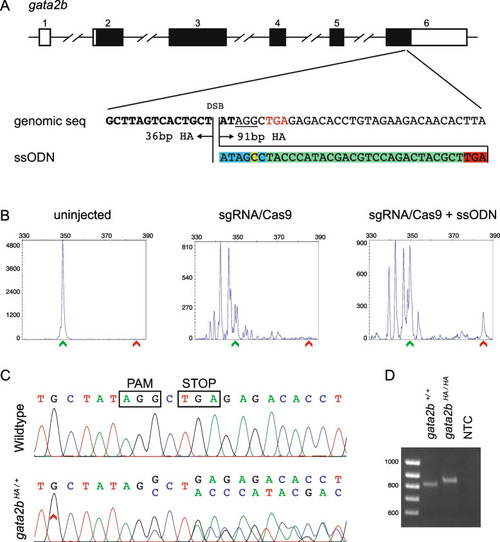
Design, screening, and validation of knock-in of HA tag at 3’ end of gata2b. A Schematic of gata2b genomic structure with coding region shaded in black (top panel), alignment of genomic sequence and ssODN template with direction of 36 bp and 91 bp homology arms (HA) marked. The ssODN contains a spacer (highlighted in blue) to maintain the ORF of gata2b as the double stranded break occurs 6 bp upstream of the stop codon (TGA, marked in red), a modification to the PAM site (G > C, highlighted in yellow) to prevent recutting after integration and the HA tag to be inserted (highlighted in green). B Representative CRISPR-STAT plots of uninjected, sgRNA/Cas9, and sgRNA/Cas9 plus ssODN injected embryos with X-axis showing the size of the peaks, Y-axis showing the peak height, and the size of the expected WT allele denoted by a green arrowhead. These plots were quantified for the presence of a peak generated by the insertion of ssODN (denoted by red arrowhead). C Sequence chromatograms showing WT and a positive F1 embryo to confirm germline transmission of knock-in sequence. D An agarose gel image showing expression of HA tag at gata2b locus by RT-PCR. RNA from gata2b+/+ or gata2bHA/HA embryos were used as template and water was used as the no template control (NTC). The original gel image is provided in additional file 2: Fig. 2 – Supporting data and its cropped version is shown here |
|
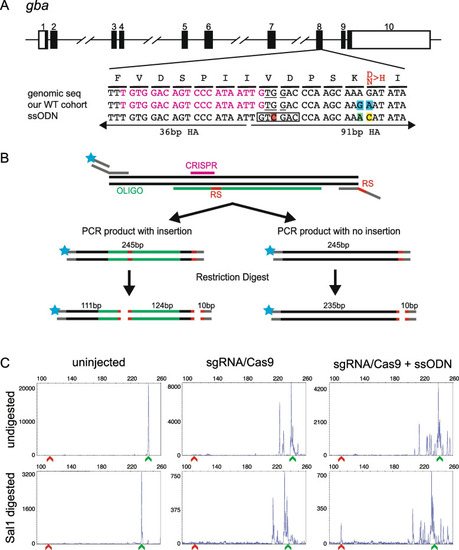
Design of ssODN and screening strategy for knock-in of a point mutation in gba. A Schematic of gba genomic structure with coding region shaded in black (top panel), alignment of genomic sequence, sequence in our cohort of fish showing 2 polymorphisms (highlighted in blue) and ssODN design with direction of 36 bp and 91 bp homology arms (HA) marked. Amino acids coded by each triplet are marked above the genomic sequence, with the amino acid to be changed marked in red. The ssODN contains a modification to the PAM site (G > C, highlighted in red) which creates a SalI restriction site (marked by black rectangle), a G > A modification to change the SNP in our WT cohort back to the reference sequence (highlighted in green) and the desired point mutation A > C (highlighted in yellow). B A schematic of the strategy for combining fluorescent PCR and RFLP analysis to detect integration of ssODN. Top panel shows the 3-primer fluorescent PCR strategy with fluorophore denoted by blue star and restriction sites marked as RS. Middle panel shows the two possible outcomes in injected embryos with size of the PCR product and location of either 2 (insertion of ssODN) or 1 (no insertion of ssODN) restriction sites (marked in red). The bottom panel shows the expected fragment sizes after the enzymatic digestion of the PCR product. Since only the fluorescently labelled fragments will be detected, embryos with knock-in can be identified by the presence of a 111 bp fragment. Multiple peaks around the WT size of 235 bp are expected due to CRISPR-induced random indels. C Representative CRISPR-STAT plots of uninjected, sgRNA/Cas9, and sgRNA/Cas9 plus ssODN injected embryos before and after SalI digestion with X-axis showing the size of the peaks, Y-axis showing the peak height, and the size of the expected WT allele denoted by a green arrowhead. Successful integration following digest results in a peak at 111 bp (denoted by red arrowhead) |
|
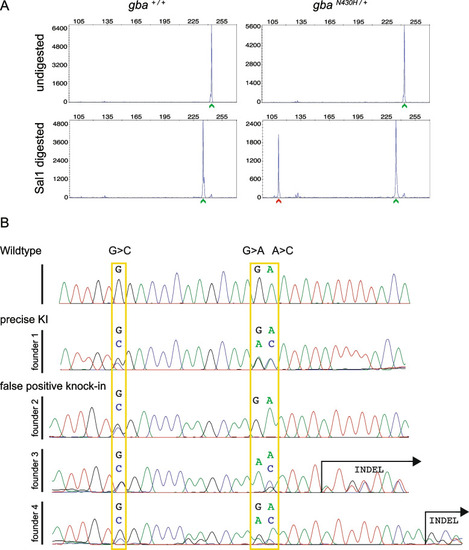
Germline screening data for knock-in of the point mutation in gba. A Representative plots of undigested and SalI digested fluorescent PCR products showing detection of the mutant allele (N430H) denoted by the red arrowhead in the progeny of a germline transmitting founder. In all plots, the X-axis represents the size of the peaks and the Y-axis shows the peak height. WT alleles (denoted by green arrowhead) are slightly smaller after digest (bottom panel) due to the restriction site in reverse primer used as internal control for successful restriction enzyme digestion. B Sequence chromatograms from representative embryos from each of the 4 founders positive for somatic knock-in compared with a WT embryo. Expected nucleotide modifications by knock-in of the ssODN are marked by yellow rectangles. Only founder 1 transmitted precise integration of the ssODN with all 3 modifications while the other 3 founders were false positives. Founder 2 had incomplete integration (the nucleotide for the restriction site was modified (G > C) but the desired (A > C) allele not changed) and founders 3 and 4 had indels |
|
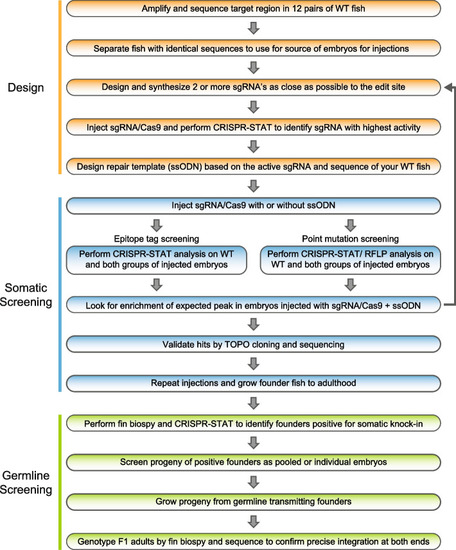
Workflow of our 3-phase knock-in pipeline. The design phase consists of five steps involving selection of WT fish for injections by sequence analysis of the target region in your WT fish line, finding an active sgRNA and designing the ssODN based on these data. The somatic screening phase occurs following injections. Embryos are collected and CRISPR-STAT or CRISPR-STAT/RFLP analysis is performed to determine if there is an enrichment of the expected peak in the sgRNA/Cas9 + ssODN group. If no enrichment is seen, another sgRNA/ssODN combination needs to be designed and tested. If expected size peaks are observed, representative samples are TOPO cloned and sequenced to confirm knock-in. The last phase is germline screening and begins by pre-screening of adult founder fish by fin biopsies for somatic knock-in. Positive fish are then prioritized for breeding to screen for germline transmission. Founders that transmit knock-in allele to their progeny are bred to grow F1 adults for genotyping, and sequence confirmation of precise knock-in |