Fig. 1
- ID
- ZDB-FIG-221214-59
- Publication
- Carrington et al., 2022 - A robust pipeline for efficient knock-in of point mutations and epitope tags in zebrafish using fluorescent PCR based screening
- Other Figures
- All Figure Page
- Back to All Figure Page
|
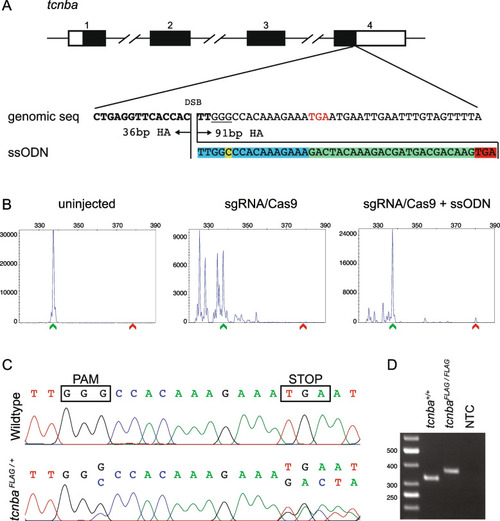
Design, screening, and validation of knock-in of FLAG tag at 3’ end of tcnba. A Schematic of tcnba genomic structure with coding region shaded in black (top panel), alignment of genomic sequence and ssODN template with direction of 36 bp and 91 bp homology arms (HA) marked. The ssODN contains a spacer (highlighted in blue) to maintain the ORF of tcnba as the double stranded break occurs 16 bp upstream of the stop codon (TGA, marked in red), a modification to the PAM site (G > C, highlighted in yellow) to prevent recutting after integration and the FLAG tag to be inserted (highlighted in green). B Representative CRISPR-STAT plots of uninjected, sgRNA/Cas9, and sgRNA/Cas9 plus ssODN injected embryos with X-axis showing the size of the peaks, Y-axis showing the peak height, and the size of the expected WT allele denoted by a green arrowhead. These plots were quantified for the presence of a peak generated by the insertion of ssODN (denoted by red arrowhead). C Sequence chromatograms showing WT and a positive F1 embryo to confirm germline transmission of knock-in sequence. D An agarose gel image showing expression of the FLAG tag at tcnba locus by RT-PCR. RNA from tcnba+/+ or tcnbaFLAG/FLAG embryos were used as template and water was used as the no template control (NTC). The original gel image is provided in additional file 2: Fig. 1 – Supporting data and its cropped version is shown here |
| Gene: | |
|---|---|
| Fish: | |
| Anatomical Term: | |
| Stage: | Day 5 |