Fig. 4
- ID
- ZDB-FIG-221011-9
- Publication
- Vandestadt et al., 2021 - RNA-induced inflammation and migration of precursor neurons initiates neuronal circuit regeneration in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
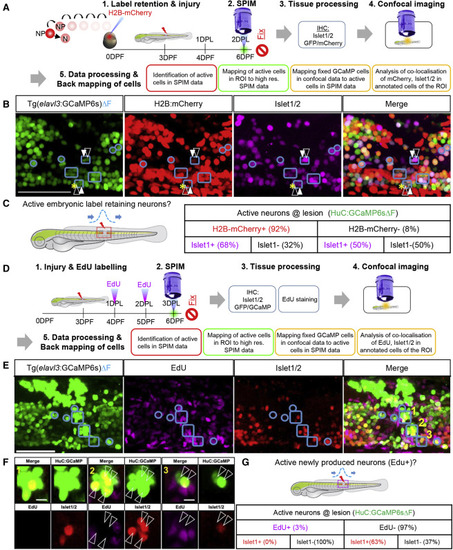
Figure 4. Role of pre-existing versus newly produced neurons in pioneering local circuit regeneration after spinal cord injury (A) Experimental design of a label retention assay to identify the contribution of embryonically produced neurons in pioneering circuit function. elavl3:H2B-GCaMP6s embryos were injected with H2B-mCherry mRNA. Neurons produced during development (N) retain mCherry expression, whereas neural progenitors (NP) that proliferate dilute the label with each round of division. At 2 DPL, larvae were imaged to identify actively firing neurons (ΔF) at the lesion site. Additionally, high-resolution images were taken, followed by fixation, Islet1/2 immunostaining, and confocal imaging. The active neurons were identified by defining active regions of interest (ROIs) within the lesion site and correlating the data to confocal data with mCherry+ cells (label retention) and Islet1/2+ cells (post-mitotic ventral marker). (B) Mapping of ΔF active ROIs (outlined in blue) to define the identity of actively firing cells at the lesion site at 2 DPL. Active ROIs may contain H2B-mCherry+ or H2B-mCherry− cells (white-filled arrowheads and white-outlined arrowheads, respectively). (C) Quantification of different cell types within the ΔF active ROIs (details in Figure S4) showing that the majority of the active neurons at the lesion were produced in the embryo. (D) Experimental design to identify the role of newly produced neurons after injury. Following injury at 3 dpf, elavl3:H2B-GCaMP6s fish were pulsed with EdU at 1 and 2 DPL. At 3 DPL, larvae were imaged to acquire GCaMP6sΔF data and high-resolution images, then fixed for staining. (E) ΔF active ROIs at the lesion were mapped onto the confocal data (outlined in blue) with EdU-labeled cells (newly produced cells) and Islet1/2 immunostaining. Active ROI cell clusters may contain cells that are EdU+ or EdU− (white-filled arrowheads and white-outlined arrowheads, respectively). The merge panel shows GCaMP, Islet1/2, and EdU cells. Yellow numbers refer to images in (F). (F) High magnification of ΔF active ROIs in E showing that active neurons do not co-localize with EdU+ cells (white open arrow heads) or Islet1/2+ cells. (G) Quantification of marker expression within ΔF active ROIs (details in Figure S4) showing that newly produced cells very rarely contribute to the functional circuitry at 3 DPL. Scale bar, 50μm. |
Reprinted from Developmental Cell, 56, Vandestadt, C., Vanwalleghem, G.C., Khabooshan, M.A., Douek, A.M., Castillo, H.A., Li, M., Schulze, K., Don, E., Stamatis, S.A., Ratnadiwakara, M., Änkö, M.L., Scott, E.K., Kaslin, J., RNA-induced inflammation and migration of precursor neurons initiates neuronal circuit regeneration in zebrafish, 2364-2380.e8, Copyright (2021) with permission from Elsevier. Full text @ Dev. Cell