Fig. 2
- ID
- ZDB-FIG-220701-26
- Publication
- Kenny et al., 2022 - TFAP2 paralogs facilitate chromatin access for MITF at pigmentation and cell proliferation genes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
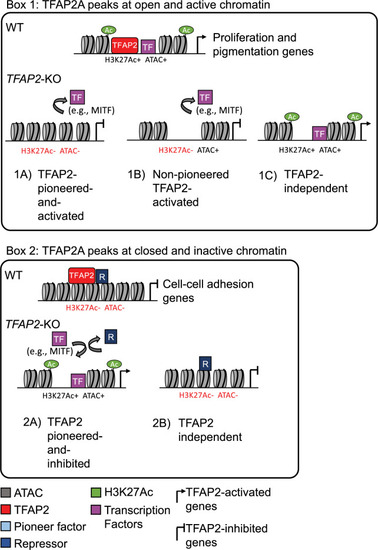
Categories of TFAP2-regulated enhancers.
Box 1: TFAP2A peaks at open and active chromatin. (1A) TFAP2-pioneered-and-activated enhancers show reduced nucleosome accessibility (ATAC-seq) and reduced levels of active chromatin marks (H3K27Ac and H3K4Me3) in TFAP2-KO cells compared to in WT cells. We infer that TFAP2 paralogs pioneer chromatin access for transcriptional co-activators, like MITF and SOX10 (purple box), that in turn recruit chromatin remodelling enzymes and histone modifying enzymes. (1B) Non-pioneered TFAP2-activated enhancers show loss of active chromatin marks but unchanged nucleosome accessibility in TFAP2-KO cells compared to in WT cells. At these enhancers, we infer that TFAP2 paralogs recruit the binding of transcription factors that, in turn, recruit histone modifying enzymes. TFAP2 paralogs also may recruit such enzymes. It is possible that these elements are stably pioneered by TFAP2 paralogs 94. (1C) At TFAP2-independent elements, neither the nucleosome accessibility nor active histone marks are altered in TFAP2-KO cells relative to in WT cells. Both types of TFAP2-activated enhancer are significantly enriched near genes whose expression is reduced in TFAP2-KO cells relative to in WT cells (i.e., TFAP2-activated genes). Such genes are associated with the gene ontology (GO) terms cell proliferation and pigmentation. TFAP2-independent elements are associated with neither TFAP2-activated nor TFAP2-inhibited genes. Box 2: TFAP2A peaks at closed and inactive chromatin. (2A) TFAP2-pioneered-and-inhibited enhancers show increased nucleosome accessibility and increased levels active chromatin marks in TFAP2-KO cells compared to in WT cells. At these sites we infer that TFAP2 paralogs recruit or stabilize the binding of enzymes that condense chromatin and that inhibit the binding of transcriptional activators that are otherwise inclined to bind at them. These elements are significantly enriched near genes whose expression is elevated in TFAP2-KO cells relative to in WT cells (i.e., TFAP2-inhibited genes). Such genes were associated with the GO terms cell-cell adhesion and cell migration. (2B) At TFAP2-independent elements, neither the nucleosome accessibility nor active histone marks are altered in TFAP2-KO cells relative to in WT cells. These elements were associated with neither TFAP2-activated nor TFAP2-inhibited genes. |
| Fish: | |
|---|---|
| Observed In: | |
| Stage Range: | Prim-5 to Prim-25 |