Fig. 2
- ID
- ZDB-FIG-211009-14
- Publication
- Jang et al., 2021 - Epigenetic dynamics shaping melanophore and iridophore cell fate in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
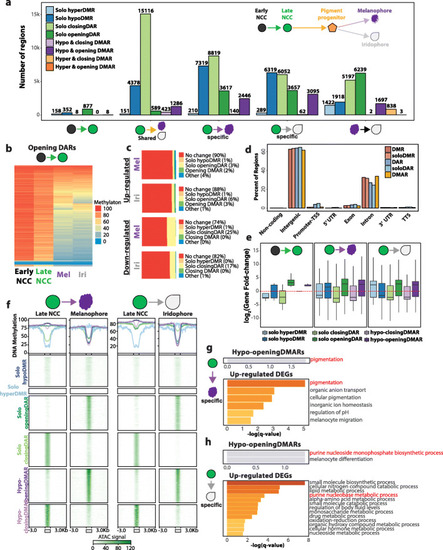
Characterization and annotation of DMARs. a Bar plot illustrating the number of DMARs identified across pigment cell differentiation. No HyperDMARs were detected, except in melanophore vs. iridophore comparison. b Heatmap illustrating the DNA methylation levels of opening DARs identified in early NCC to late NCC transition. c Epigenetic dynamics of DEG promoters in melanophores and iridophores. d Genomic feature distribution of DMRs, DARs, and DMARs. e Expression fold-change of closest DEGs within 50 kb of epigenetically dynamic regions. f Line graphs and heatmaps representing average DNA methylation levels and ATAC peak signals respectively of epigenetically dynamic regions from late NCC to pigment cell-type comparison. g, h Gene ontology enrichment of DEGs within 50 kb of hypo-opening DMARs and upregulated DEGs in melanophore-specific (g) and iridophore-specific (h) comparison |