Figure 1
- ID
- ZDB-FIG-210915-86
- Publication
- de Vrieze et al., 2021 - Efficient Generation of Knock-In Zebrafish Models for Inherited Disorders Using CRISPR-Cas9 Ribonucleoprotein Complexes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
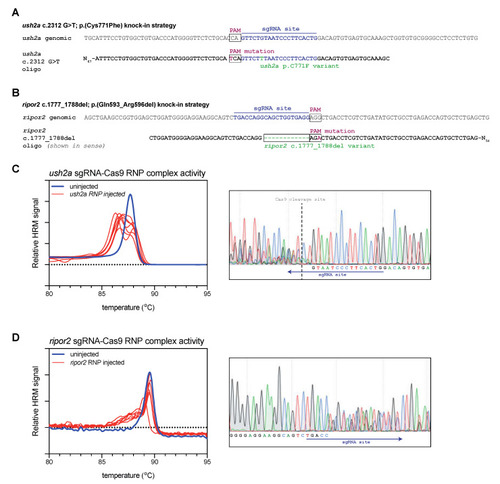
Study design and validation of sgRNA-Cas9 ribonucleoprotein complex efficiency. (A) Graphic representation of the target region in ush2a exon 13. The sgRNA (targeting the complementary strand) and protospacer-adjacent motif (PAM) are indicated and allow for Cas9 cleavage in close proximity of position c.2312. The asymmetric antisense template for homology directed repair contains the c.2312G>T variant and a silent PAM-site change. (B) Graphic representation of the target region in ripor2 exon 14. The sgRNA is designed to direct Cas9 to cleave within the nucleotide sequence that we aimed to delete. The asymmetric antisense template for homology directed repair, containing the 12-nucleotide deletion and a silent PAM-site change, is shown in sense orientation. (C) High resolution melting (HRM) analysis profiles of individual embryos injected with the ush2a sgRNA-Cas9 ribonucleoprotein (RNP) complex. A representative example of the Sanger sequencing traces from a single embryo is shown on the right. The typical overlapping peaks occur around the indicated Cas9 cleavage site. (D) HRM analysis profiles of individual ripor2 sgRNA-Cas9 ribonucleoprotein (RNP) complex injected embryos. The typical overlapping peaks in the sanger sequencing traces, shown on the right, are indicative of sgRNA-Cas9 RNP activity. |