Fig. 4
- ID
- ZDB-FIG-210413-44
- Publication
- Dietrich et al., 2021 - Skeletal biology and disease modeling in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
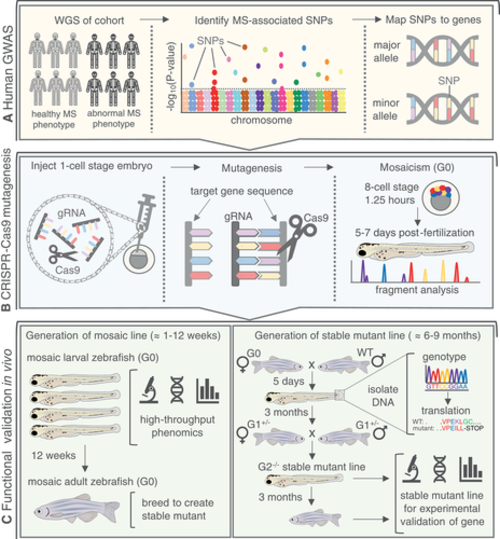
Zebrafish as a tool for rapid validation of GWAS‐derived MS disease‐associated gene candidates. (A) Human GWAS conduct WGS on large cohorts and identify SNP mutations in people with abnormal MS phenotypes. SNPs are then mapped to the nearest gene using whole‐exome sequencing. (B) CRISPR‐Cas9 technology can be used for targeted knockout mutagenesis in vivo. Gene‐specific gRNAs are designed and injected into a one‐cell stage zebrafish embryo with Cas9. Efficient gRNAs will facilitate double‐strand breaks within an exon in the target gene, resulting in indel mutations. A mosaic (G0) embryo (crispant) will develop, containing a variety of mutant and WT alleles for the gene of interest that are validated through fragment analysis. (C) (Left) mosaic larval zebrafish can be rapidly screened for MS phenotypes during skeletogenesis, providing a high‐throughput and efficient phenomics‐based approach to gene validation. Mosaic G0 zebrafish can be raised to adulthood for breeding into a stable line with a single, known mutant allele. (Right) G0 mosaic zebrafish are crossed to WT. Resulting heterozygous larvae (G1+/−) are fin‐clipped to isolate DNA for genotyping and in silico translation to identify alleles resulting in a premature STOP codon, compromising the protein. G1+/− with the same mutant allele are bred with each other to generate the G2−/− line with a stable mutation in the gene of interest. The stable mutant larvae or adults can then be used for experimental validation of the MS‐associated phenotype. gRNA = guide RNA; hpf = hours post fertilization; MS = musculoskeletal; SNP = single‐nucleotide polymorphism; WGS = whole‐genome sequencing; WT = wild‐type. |