Figure 8
- ID
- ZDB-FIG-210307-103
- Publication
- Li et al., 2021 - Prpf31 is essential for the survival and differentiation of retinal progenitor cells by modulating alternative splicing
- Other Figures
- All Figure Page
- Back to All Figure Page
|
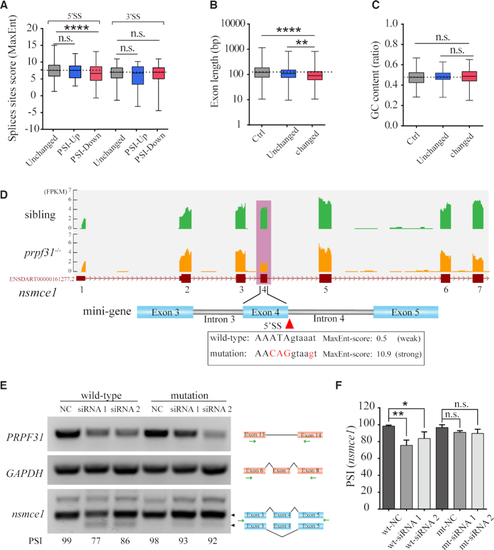
Prpf31 deletion is more likely to cause the skipping of exons with shorter length and weaker 5′ splicing site. ( |