|
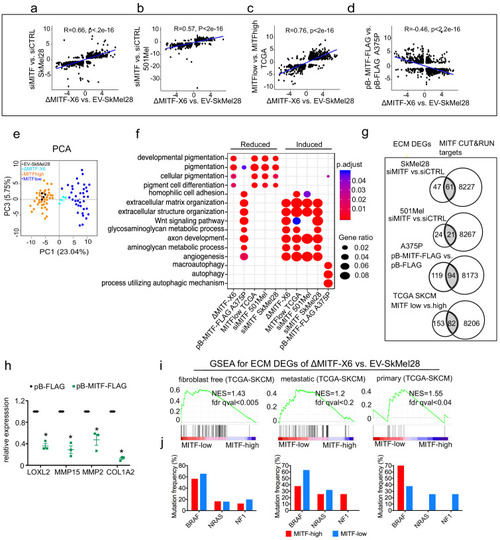
The extracellular matrix (ECM) and focal adhesion gene signature is overrepresented upon MITF depletion and in MITF<sup>low</sup> human melanoma tumors.(a–d) Positive correlation of DEGs in ∆MITF-X6 vs. EV-SkMel28 cells with DEGs of siMITF vs. siCTRL in 501Mel and SkMel28 and MITFlow vs. MITFhigh melanoma tumors from TCGA, and negative correlation of DEGs in pB-FLAG vs. pB-MITF-FLAG A375P cells is shown. Values used in the X and Y axis are log2 fold change in the expression of DEGs. (e) Principal component analysis (PCA) of the 200 most significant DEGs in the MITFlow vs. MITFhigh and ∆MITF-X6 vs. EV-SkMel28 display similar clustering where EV-SkMel28 samples cluster with MITFhigh tumors and ∆MITF-X6 cells with MITFlow tumors. (f) GO BP analysis of induced and reduced DEGs affected by MITF KO/KD in SkMel28 and 501Mel cells, and DEGs affected by MITF overexpression in A375P cells. (g) Venn diagram displaying the overlap in the number of differentially expressed ECM genes affected by MITF and MITF CUT-and-RUN targets. (h) RT-qPCR showing reduced expression of ECM genes in the stable dox-inducible MITF overexpression A375P cell line (pB-MITF-FLAG). Relative expression was calculated by normalizing to control cells expressing empty vector (pB-FLAG). Error bars indicate standard error of the mean (* p value <0.05) was calculated using paired t-test. (i) Gene set enrichment analysis using ECM genes differentially expressed between ∆MITF-X6 and EV-SkMel28 cells in the top 30 MITFlow and 30 MITFhigh samples with high fibroblast marker removed Primary, and metastatic melanoma from TCGA were analyzed separately. (j) Percentage of mutations in the indicated genes in the MITFlow and MITFhigh tumors from fibroblast-free, primary and metastatic TCGA tumor samples, respectively.
|