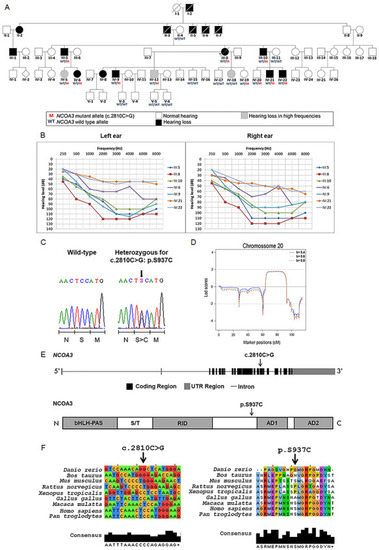
A rare variant in NCOA3, a gene which codes a nuclear receptor coactivator, segregates with hearing loss in the family. A) Pedigree showing the segregation of the NCOA3 variant (NM_181659: c.2810C > G: p.Ser937Cys). B) 14 audiometric profiles (divided in right and left ear) of 7 patients affected with sensorineural and bilateral hearing loss. Hearing thresholds until 20 dB are considered normal. C) Chromatograms showing partial sequence from affected patient compared to a wild-type sequence. Arrow indicates position of the NCOA3 variant, while scale bar below indicates which amino acid is changed when the variant is present. D) Multipoint LOD scores calculated with Merlin software for chromosome 20, using data from SNP arrays, under assumption of penetrance K = 0.4, K = 0.6 and K = 0.8. E) Schematics of NCOA3 gene and its respective protein. bHLH = basic helix–loop–helix domain; PAS = Per/ARNT/Sim homologous domain; S/T = serine/threonine-rich region; RID = receptor interaction domain containing multiple LXXLL motifs; AD1 and AD2 = activation domains 1 and 2. F) Multiple alignment of NCOA3 gene and its orthologous (left), as well as multiple alignment of the respective proteins (right). Arrow indicates position of NCOA3 variant (NM_181659: c.2810C > G: p.Ser937Cys).
|