Figure 1
- ID
- ZDB-FIG-210123-23
- Publication
- Almeida et al., 2021 - Endogenous zebrafish proneural Cre drivers generated by CRISPR/Cas9 short homology directed targeted integration
- Other Figures
- All Figure Page
- Back to All Figure Page
|
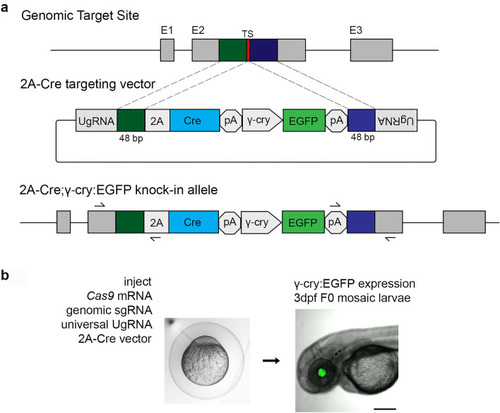
CRISPR/Cas9 short homology directed targeted integration strategy for efficient recovery of Cre knock-in alleles. ( |