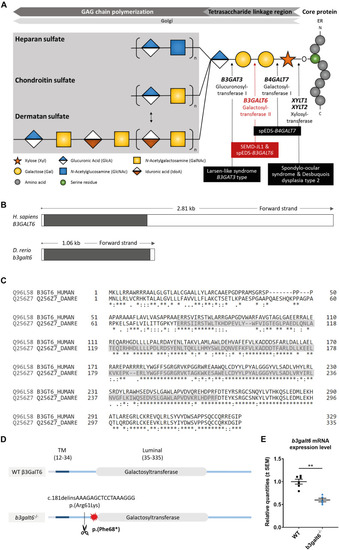
Schematic representation of the GAG biosynthesis and the evolutionary conservation of the zebrafish and human b3galt6/B3GALT6 gene and B3galt6/β3GalT6 protein. (A) Simplified schematic representation of the PG linkage and GAGs biosynthesis. PG core proteins are synthesized in the endoplasmic reticulum and undergo further modification in the Golgi apparatus. First, a tetrasaccharide linkage region is synthesized that originates from the addition of a Xyl onto a serine residue of the core protein. Subsequent addition of two Gals and one GlcA residues completes this process. Depending on the addition of GlcNAc or GalNAc to the terminal GlcA residue of the linkage region, HS or CS/DS PGs will be formed, respectively. The elongation of the GAG chain continues by repeated addition of uronic acid and amino sugar residues. The GAG chain is then further modified by epimerization and sulfation (not shown). The main genes encoding for the enzymes involved in the linkage region biosynthesis are indicated in bold and biallelic mutations in these genes result in different linkeropathies. Disorders resulting from deficiency of the enzymes are indicated in black boxes. The deficient enzyme (β3GalT6) and the related disorder studied here are depicted in red (e.g., spEDS-B3GALT6 and SEMD-JL1). (B) Schematic representation of the genomic structure of the H. sapiens B3GALT6 (NM_080605.4) and D. rerio b3galt6 (NM_001045225.1) genes. The white and black boxes represent the untranslated regions and the single B3GALT6/b3galt6 exon, respectively. The zebrafish b3galt6 gene resides on chromosome 11, spans a region of 1.06 kb and has not been duplicated during teleost evolution. (C) Amino acid (AA) sequence alignment (Clustal Omega) between H. sapiens β3GalT6 (Uniprot: Q96L58) and D. rerio B3galt6 (Uniprot: Q256Z7). The luminal galactosyltransferase domain is highlighted in gray. Conservation of AA sequences is shown below the alignment: “*” residues identical in all sequences in the alignment; “:” conserved substitutions; “.” semi-conserved substitutions; “space” no conservation. (D) Schematic representation of the cmg20 allele in b3galt6 mutants. b3galt6 consists of three parts, a cytosolic domain (AA 1-11), a transmembrane domain (TM, AA 12-34) and a luminal domain (AA 35-335). The location of the CRISPR/Cas9-generated indel variant is indicated by a scissor symbol and the resulting premature termination codon (PTC) is depicted by a red star. (E) RT-qPCR analysis of relative b3galt6 mRNA expression levels in b3galt6–/– adult zebrafish (0.597 ± 0.03919) (n = 5) compared to WT siblings (1 ± 0.05395) (n = 6). Data are expressed as mean ± SEM. A Mann-Whitney U test was used to determine significance, **P < 0.01.
|

