Fig. 1
- ID
- ZDB-FIG-201013-10
- Publication
- Kushawah et al., 2020 - CRISPR-Cas13d Induces Efficient mRNA Knockdown in Animal Embryos
- Other Figures
- All Figure Page
- Back to All Figure Page
|
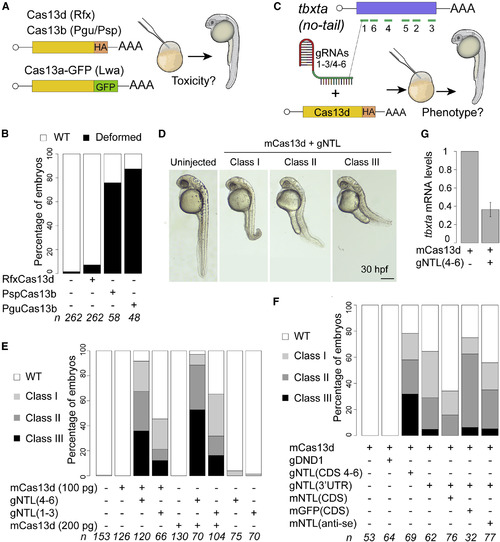
CRISPR-RfxCas13d System Targeting tbxta Recapitulates No-Tail Phenotype in Zebrafish Embryos (A) Schematic illustration of the experimental setup used to analyze CRISPR-Cas13 toxicity. 200 pg of mRNA from each Cas13 variant was injected into one-cell-stage zebrafish embryos. Cas13b and Cas13d are tagged with HA and Cas13a is fused to GFP. (B) Toxicity evaluation of embryos. Stacked barplots showing the percentage of deformed and wild-type (WT) zebrafish embryos (30 hpf) injected in conditions described in (A). Number of embryos evaluated (n) is shown for each condition. (C) Schematic of positions of six gRNAs (green lines) targeting tbxta CDS in zebrafish (top). Schematic of experimental setup to analyze CRISPR-RfxCas13d-mediated RNA knockdown in zebrafish (bottom). Two sets of gRNAs targeting tbxta CDS (1–3 or 4–6; 300 pg per embryo) were mixed with mRNA (100 or 200 pg per embryo) coding for RfxCas13d (mCas13d) and injected into one-cell-stage embryos. (D) Phenotypic severity in embryos injected with mCas13d and gRNAs targeting tbxta (gNTL) compared with uninjected WT evaluated at 30 hpf (scale bar, 0.5 mm). Class I: short tail (least extreme). Class II: absence of notochord and short tail (medium level). Class III: absence of notochord and extremely short tail (most extreme). (E) Different gNTL sets recapitulate no-tail phenotype. Stacked barplots showing percentage of observed phenotypes under various injection conditions. Number of embryos evaluated (n) is shown for each condition. (F) No-tail phenotype can be rescued. Stacked barplots showing percentage of observed phenotypes using gNTLs targeting 3′UTR (300 pg/embryo) and mCas13d (200 pg/embryo) co-injected with different mRNAs: GFP (mGFP CDS, 100 pg/embryo), tbxta ORF with 3′UTR from pSP64T plasmid (mNTL CDS, 100 pg/embryo), or tbxta antisense (mNTL anti-se, 100 pg/embryo) ORF. As an additional control, an unrelated gRNA set targeting a gene involved in germ cell formation (dnd1) was included (300 pg/embryo). Number of embryos evaluated (n) is shown for each condition. (G) qRT-PCR analysis showing levels of tbxta mRNA at 6 hpi in conditions indicated. Results are shown as the averages ± standard error of the mean from two independent experiments with 1 or 2 biological replicates per experiment (n = 20 embryos/biological replicate) for mCas13d alone and Cas13d plus gNTL, respectively. p = 0.0092 (t test). cdk2ap mRNA was used as normalization control. |
Reprinted from Developmental Cell, 54(6), Kushawah, G., Hernandez-Huertas, L., Abugattas-Nuñez Del Prado, J., Martinez-Morales, J.R., DeVore, M.L., Hassan, H., Moreno-Sanchez, I., Tomas-Gallardo, L., Diaz-Moscoso, A., Monges, D.E., Guelfo, J.R., Theune, W.C., Brannan, E.O., Wang, W., Corbin, T.J., Moran, A.M., Sánchez Alvarado, A., Málaga-Trillo, E., Takacs, C.M., Bazzini, A.A., Moreno-Mateos, M.A., CRISPR-Cas13d Induces Efficient mRNA Knockdown in Animal Embryos, 805-817.e7, Copyright (2020) with permission from Elsevier. Full text @ Dev. Cell