Fig. 9
- ID
- ZDB-FIG-200421-2
- Publication
- Stanney et al., 2019 - Combinatorial action of NF-Y and TALE at embryonic enhancers defines distinct gene expression programs during zygotic genome activation in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
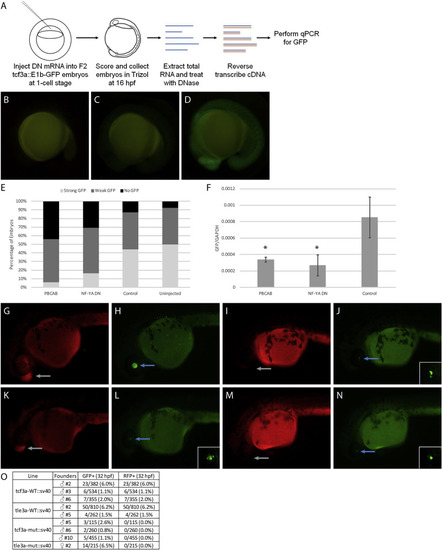
Disruption of TALE and NF–Y function reduces enhancer activity. (A) Schematic showing workflow for dominant negative disruption of tcf3a:E1b-GFP. (B–D) Representative images showing no GFP (B), weak GFP (C), and strong GFP (D) of dominant negative-injected embryos. (E) Distribution of GFP expression in uninjected embryos and embryos injected with PBCAB, NF-YA DN or control RNA. (F) RT-qPCR-based detection of GFP expression in embryos injected with PBCAB, NF-YA DN or control RNA. Data are shown as mean±SEM. Statistical test: unpaired t-test. (G–N) Representative examples of RFP (G, K, I, M) and GFP (H, L, J, N) signal in tcf3a-WT:sv40 (G, H), tcf3a-mut:sv40 (I, J), tle3a-WT:sv40 (K, L) and tle3a-mut:sv40 (M, N) embryos at 32hpf. Insets in panels L, J, N show higher magnification of GFP expression in lens. Note that embryo in panels G, H is at a later stage than embryos in panels I–N. (O) Table quantifying results from experiment in panels G–N. |
Reprinted from Developmental Biology, 459(2), Stanney, W., Ladam, F., Donaldson, I.J., Parsons, T.J., Maehr, R., Bobola, N., Sagerström, C.G., Combinatorial action of NF-Y and TALE at embryonic enhancers defines distinct gene expression programs during zygotic genome activation in zebrafish, 161-180, Copyright (2019) with permission from Elsevier. Full text @ Dev. Biol.