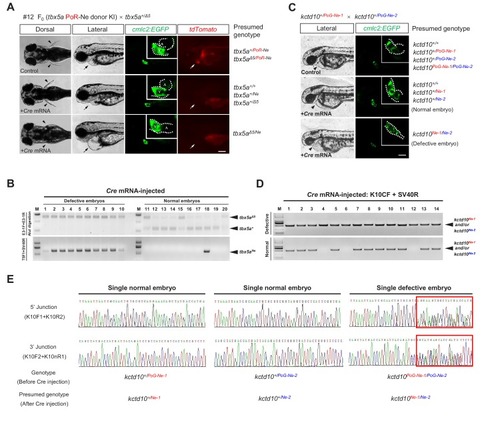
Figure 2—figure supplement 1.
- ID
- ZDB-FIG-191230-1778
- Publication
- Li et al., 2019 - One-step efficient generation of dual-function conditional knockout and geno-tagging alleles in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
( |