- Title
-
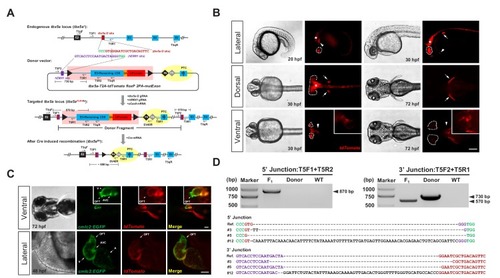
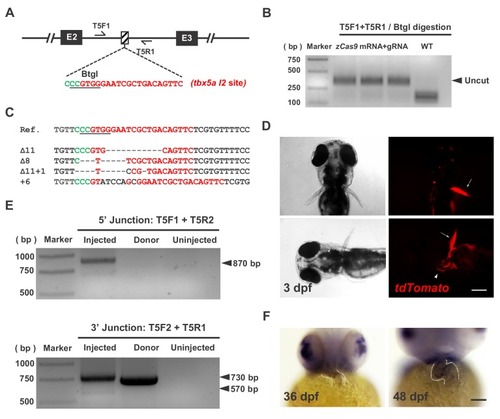
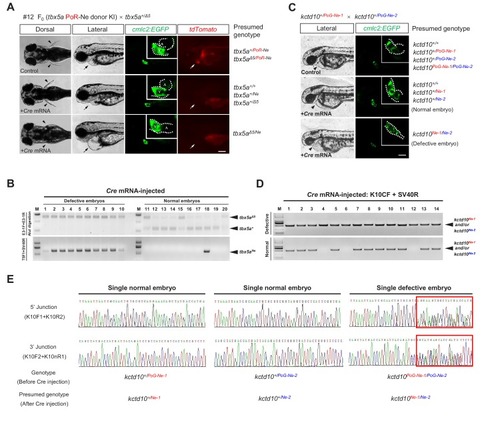
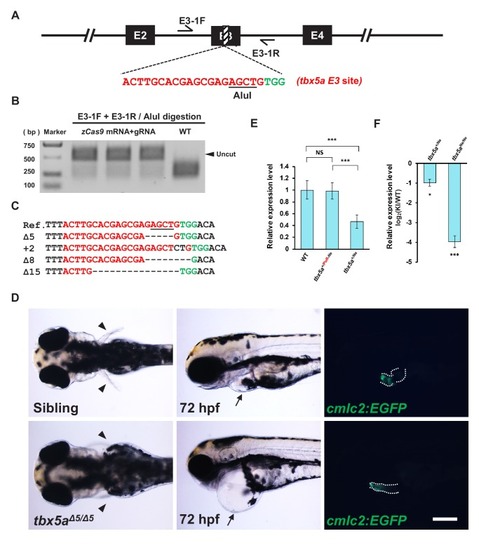
One-step efficient generation of dual-function conditional knockout and geno-tagging alleles in zebrafish
- Authors
- Li, W., Zhang, Y., Han, B., Li, L., Li, M., Lu, X., Chen, C., Lu, M., Zhang, Y., Jia, X., Zhu, Z., Tong, X., Zhang, B.
- Source
- Full text @ Elife
|
( |
|
( |
|
( |
|
( |
|
( |
|
( |
|
( |
|
( |
|
( |