Fig. 1
- ID
- ZDB-FIG-180529-27
- Publication
- Bajanca et al., 2017 - Epigenetic Regulators Modulate Muscle Damage in Duchenne Muscular Dystrophy Model
- Other Figures
- All Figure Page
- Back to All Figure Page
|
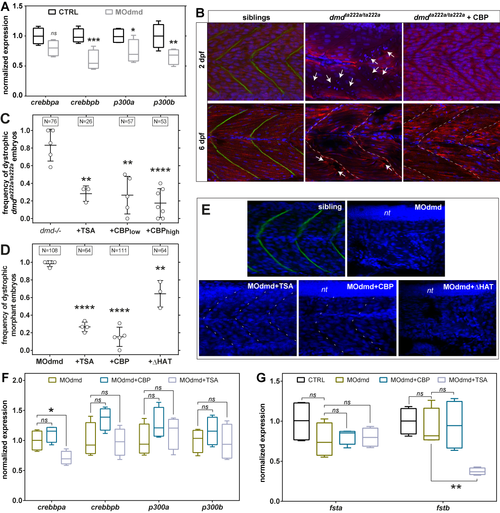
Epigenetic regulators in zebrafish dystrophic embryos. A) Quantitative PCR analysis of p300/CBP transcript expression at 2 days post fertilisation (dpf) in control and morpholino-depleted embryos (MOdmd). (B) Confocal sections of Tg(actc1b:mCherry)pc4 embryos immunostained for Dystrophin (green). Well organised pattern of muscle fibres (mCherry, red) and nuclei (DAPI, blue) in siblings, while heavily disrupted in dmdta222a/ta222a embryos (arrows), showing collapsed and wavy fibres which ends misalign with somatic borders (dashed lines). Overexpressing CBP (dmdta222a/ta222a + CBP; injection of 107 RNA molecules of plasmid per embryo at one cell stage) has a clear positive effect on dystrophic embryos both at 2 and 6 dpf. (C, D) Frequency of dystrophic phenotype (see Materials and Methods) on 2 dpf Dystrophin-null embryos, either dmdta222a/ta222amutants (C) or depleted with MOdmd (D). Overexpressing CBP (low = 10 |
| Genes: | |
|---|---|
| Antibody: | |
| Fish: | |
| Condition: | |
| Knockdown Reagents: | |
| Anatomical Terms: | |
| Stage Range: | Long-pec to Day 6 |
| Fish: | |
|---|---|
| Condition: | |
| Knockdown Reagents: | |
| Observed In: | |
| Stage Range: | Long-pec to Day 6 |