Fig. 3
|
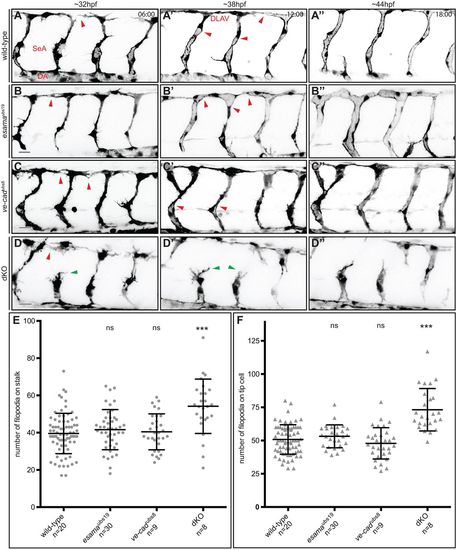
Loss of both VE-cad and Esama enhances the defects observed in ve-cadubs8 mutants. (A-D) Still images from Movie 4 (A), Movie 5 (B), Movie 6 (C) and Movie 7 (D) of Tg(fli1a:EGFP)y1 wild-type, esamaubs19, ve-cadubs8 and esamaubs19; ve-cadubs8 double mutant (dKO) embryos, respectively. Images are shown in inversed contrast at three different stages of angiogenesis: ∼32, ∼38 and ∼44 hpf. Red arrowheads at 32 hpf point to anastomotic contacts; red arrowheads at 38 hpf point to forming lumen; green arrowheads point to stalk cells that detached from the tip cell. Scale bars: 20 µm. (E and F) Quantification of filopodial protrusions on SeA stalks (E) and tip cells (F). (E) The number of filopodia on angiogenic stalks (composed of one to three ECs) is comparable between wild-type (77 SeAs in n=20 embryos), esamaubs19 (114 SeAs in n=30 embryos) and ve-cadubs8 (31 SeAs in n=9 embryos) genotypes. Significantly more filopodia were counted on dKO stalks (31 SeAs in n=8 embryos). (F) Tip cells generally generate more filopodial protrusions (average of 50 filopodia per tip cell) than stalk cells. Significantly more filopodia were counted on tip cells of dKO compared with wild type. Bars in the plots represent mean±s.d. DA, dorsal aorta; dKO, double mutant; DLAV, dorsal longitudinal anastomotic vessel; n, number of analyzed embryos; ns, not significant; SA, segmental artery. ***P<0.0001, with one-way analysis of variance (ANOVA). |