Fig. 6
- ID
- ZDB-FIG-131031-19
- Publication
- Jao et al., 2013 - Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system
- Other Figures
- All Figure Page
- Back to All Figure Page
|
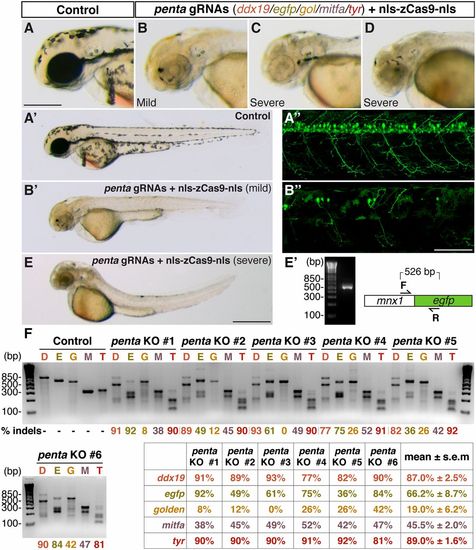
Multiplex genome editing in zebrafish by Cas9. A mix of five gRNAs (penta gRNAs: ddx19 gRNA, 55 pg; egfp gRNA, 15 pg; gol gRNA, 25 pg; mitfa gRNA, 60 pg; and tyr gRNA, 25 pg) were coinjected with nls-zCas9-nls RNA (150 pg) into Tg(-5.1mnx1:egfp) transgenic embryos. (A–E) Lateral views of the wild-type (A–A22) and the penta gRNAs/nls-zCas9-nls RNA-injected (B–B22 and C–E) embryos. The combination of three distinct phenotypes—brain necrosis/small eyes/curved body axis, hypopigmentation, and loss of EGFP expression in the motoneurons—caused by biallelic inactivations of multiple target genes simultaneously were observed in the same embryos (B–B22 and E and E2). Some quintuple knockout embryos showed no EGFP-positive motoneurons due to the almost nonmosaic disruption of the Tg(-5.1mnx1:egfp) transgene, whose presence was confirmed by PCR (E2). (F) T7EI assays showed high mutagenesis rates at five genomic targets in six randomly selected quintuple knockout embryos (penta KO 1–6: D, ddx19; E, egfp; G, gol; M, mitfa; T, tyr). (Scale bars: in A, 300 μm; in B22, 50 μm; in E, 500 μm.) |