Fig. 5
- ID
- ZDB-FIG-131031-18
- Publication
- Jao et al., 2013 - Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system
- Other Figures
- All Figure Page
- Back to All Figure Page
|
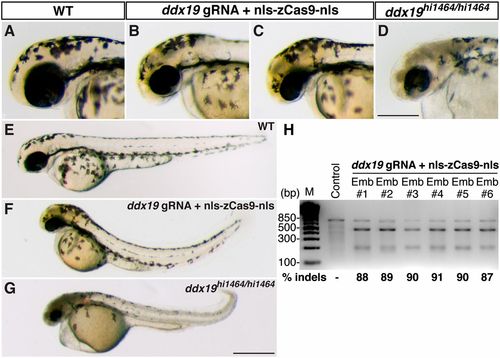
Biallelic disruption of ddx19 by Cas9 generates null-like phenotypes. ddx19 gRNA (150 pg) and nls-zCas9-nls RNA (150 pg) were injected into wild-type embryos. (A–G) Lateral views of the wild-type (A and E), ddx19-targeted (B, C, and F), and homozygous ddx19 mutants (ddx19hi1464/hi1464 ) (D and G) embryos. ddx19-targeted embryos recapitulated several ddx19 null phenotypes (e.g., brain necrosis, small eyes, and curved body axis), albeit to a lesser extent. (H) T7EI assays showed high mutagenesis rates at the ddx19 target (87–91%) in six randomly selected ddx19-targeted embryos. (Scale bars: in D, 300 μm; in G, 500 μm.) |
| Fish: | |
|---|---|
| Observed In: | |
| Stage: | Long-pec |