Fig. 1
- ID
- ZDB-FIG-110128-17
- Publication
- Kinkhabwala et al., 2011 - A structural and functional ground plan for neurons in the hindbrain of zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
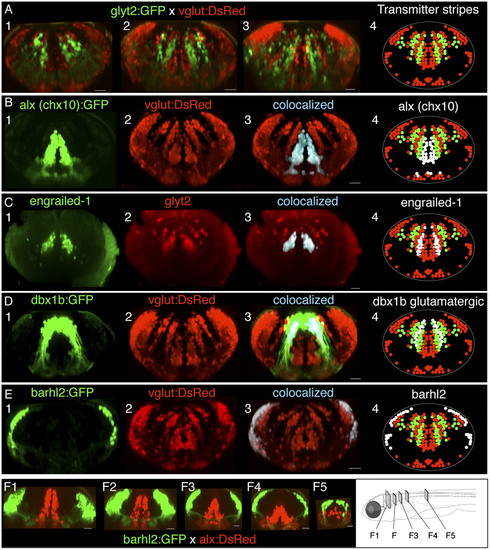
Interleaved transmitter stripes of neurons in hindbrain and their overlap with transcription factor expression patterns. (A) Cross-sections of hindbrain in vivo in rhombomeres 4, 6, and 8 (A, panels 1–3, respectively) from a glyt2:GFP x vglut:DsRed four dpf fish, showing interleaved stripes of glycinergic (green) and glutamatergic neurons (red). (B–E) Transcription factor stripes are present in hindbrain and overlap glutamatergic and glycinergic stripes. (B) Cross-sections from rhombomere 8 of a dual transgenic alx (chx10):GFP and vglut: DsRed three dpf fish shows that alx expression colocalizes with the most medial glutamatergic stripe. (C) Engrailed-1 immunostaining in cross-sections from a glyt2 transgenic line from rhombomere 7 in a 5-dpf fish showing that all medial glycinergic stripe neurons express engrailed-1. (D) Cross-section from rhombomere 7 in a four dpf vglut:DsRed x dbx1b:GFP transgenic fish shows that dbx colocalizes with both the middle glutamatergic stripe as well as a more lateral stripe in a region known to contain glycinergic neurons. (E) Cross-sections from rhombomere 8 in a 4-dpf vglut:DsRed × barhl2:GFP fish indicate that barhl2 staining overlaps the lateral portion of the most lateral glutamatergic populations. (F) Cross-sections of the transformation of transcription factor stripes from hindbrain (F1) to spinal cord (F5) in a cross of barhl2GFP and alx:DsRed lines. White regions in panel 4 in B–E indicate the location of the transcription factor expression relative to the neurotransmitter stripes. All cross-sections are maximum-intensity projections of 20- to 50-μm volumes from a confocal image stack of the entire hindbrain. (Scale bars, 20 μm.) |