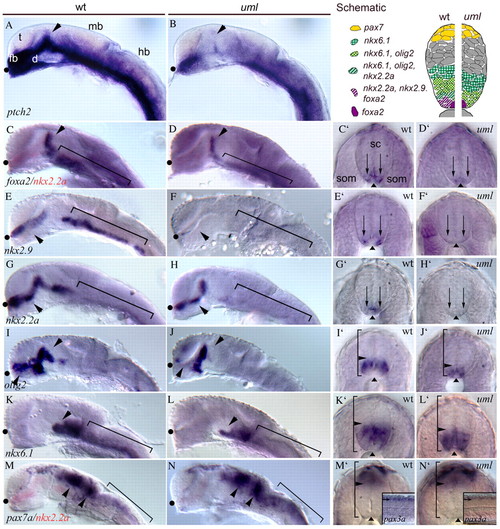
Reduced Hh signaling in uml mutants. (A) The Hh target gene ptc1 (now known as ptch2) is expressed throughout the ventral CNS in wild-type embryos with a dorsal peak of expression in the zona limitans interthalamica (ZLI) (arrowhead). (B) In uml mutants, ptch2 expression is greatly reduced throughout the brain and spinal cord, and is nearly absent from the ZLI (arrowhead). (C) foxa2 is expressed in the ZLI (arrowhead) and ventral midbrain/hindbrain (bracket). (C2) In the spinal cord, foxa2 is expressed in the medial floor plate (FP, arrowhead) and lateral FP cells that correspond to the V3 region of the mammalian spinal cord (arrows). (D,D′) In uml mutants, foxa2 expression is reduced in the brain and is absent from the lateral floor plate (D′ arrows). (E,E′) nkx2.9 is expressed in the diencephalon (arrowhead in E), ventral midbrain/hindbrain (bracket) and in lateral FP cells (arrows in E′). (F,F′) In uml mutants, nkx2.9 expression is completely absent throughout the CNS. (G,G′) nkx2.2a is expressed in the diencephalon (arrowhead in G), ventral midbrain/hindbrain (bracket) and lateral floor plate of the spinal cord (arrows in G′). (H,H′) In uml mutants nkx2.2a expression is reduced in the diencephalon (arrowhead in H), ventral midbrain/hindbrain (bracket), and spinal cord (arrows in H′). (I,I′) olig2 is regionally expressed in the diencephalon (arrowheads) and in a band of cells in the spinal cord just dorsal to the floor plate (arrowhead in I′). (J,J′) In uml mutants, olig2 expression is regionally absent in the diencephalon (arrowheads in J) and expression is reduced and shifted ventrally in the spinal cord (arrowhead in J′). (K,K′) nkx6.1 is expressed in the ventral midbrain (arrowhead in K), hindbrain (bracket in K) and in the ventral half of the spinal cord (bracket with arrowhead in K′). (L,L2) In uml mutants, nkx6.1 expression is reduced in the brain (arrowhead and bracket in L) and shifted ventrally in the spinal cord (bracket with arrowhead in L′). (M,M′) pax7 is expressed in the dorsal midbrain (arrowheads in M), hindbrain (bracket in M), and spinal cord (bracket with arrowhead in M′). Inset shows pax3 expression. (N,N′) In uml mutants, pax7 (and pax3, inset) expression appears unchanged. A-N are lateral views of the heads of 24 hpf zebrafish, anterior to the left, eyes removed. Black dots mark the optic recess. C′-N′ are spinal cord cross-sections of embryos shown in C-N. In C′-N′, arrows mark the lateral floor plate and arrowheads mark the floor plate. In I′-N′, brackets with arrowheads mark the dorsoventral boundary of gene expression within the spinal cord. In C, D, M and N nkx2.2a labeling (red) was used to identify uml mutants. Upper right panel is a schematic representation of Hh target gene expression in the spinal cord of wild-type (left) and uml mutant (right) embryos. d, diencephalon; fb, forebrain; hb, hindbrain; mb, midbrain; sc, spinal cord; som, somites; t, telencephalon.
|

