Fig. S1
- ID
- ZDB-FIG-101026-13
- Publication
- Stewart et al., 2010 - Phosphatase-Dependent and -Independent Functions of Shp2 in Neural Crest Cells Underlie LEOPARD Syndrome Pathogenesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
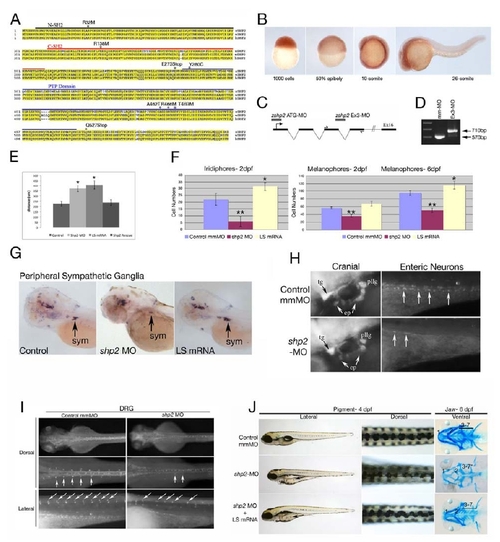
(A) Zebrafish Shp2 encodes a protein of 597 amino acids with 92% identity and 99% similarity to its murine and human orthologs. The N-SH2, C-SH2 and PTP domains are underlined black, red and blue, respectively. The locations of point mutations and truncations used in this study are indicated above the sequence. (B) Analysis of shp2 mRNA expression during embryogenesis by whole-mount in situ hybridization, showing ubiquitous maternal expression at blastula (1000-cell) and gastrulation stages. At segmentation stages, shp2 levels are highest in the developing CNS and spinal cord. (C) Schematic of shp2 genomic structure, showing the translation-blocking (ATG-MO) and splice-blocking (Ex3-MO) antisense morpholinos used in this study. The primers used to confirm interruption of splicing by Ex3-MO are indicated. (D) RT-PCR of embryos injected with Ex3-MO, showing that it prevents use of the normal shp2 exon 3 splice donor site; instead, a cryptic donor site 140 bp downstream in intron 4 is used (data not shown) and a larger PCR product results. (E) Quantification of gastrulation phenotypes represented in Figure 1A, as determined by measuring the distance from the tip of the head to the tailbud. Co-expression of human SHP2 mRNA rescues the shp2 MO gastrulation phenotypes, *p <0.005. (F) Quantification of the pigment cell defects associated with LS mRNA- and shp2 MO-injected embryos; n= 10 for all samples, *p <0.002, **p <0.0001. (G) Lateral views of peripheral sympathetic neurons (sym) detected by RNA in situ hybridization for tyrosine hydroxylase (th) at 4 dpf. Injection of the shp2 MO causes ~ 60% (p <0.002) loss of th-positive cells in the superior cervical ganglia (sym), whereas injection of LS mRNA causes a ~40% (p <0.003) increase in the number of these neurons (arrows). (H) Lateral view showing immunofluorescence labeling of sensory cranial ganglia and enteric neurons with α-Hu-C (HuC) at 4 dpf. shp2 MO-injected embryos have reduced numbers of trigeminal (tg), epibranchial (ep) and posterior lateral line (pllg) neurons and severe loss of enteric neurons (ent) neurons. (I) Dorsal root ganglia (DRG) neurons labeled with HuC at 4 dpf show loss and displacement in shp2 MO-injected embryos (arrows). (J) Co-injection of LS mRNA and shp2 MO results in LS-like NC phenotypes. Lateral (left) and dorsal (middle) views of live 4 dpf embryos, and ventral views (right) of 6 dpf embryos stained with Alcian blue. Injection of shp2 MO causes loss of pigment and jaw elements, as well as delayed migration to the ventral stripe (arrow). Co-injection of LS mRNA (A462T) with the shp2 MO (bottom) results in a LS-like pigment and jaw phenotypes in 70-80% of embryos (n=100), with delayed migration to the ventral stripe (compare with Figs. 1 D-G). Pigment numbers at 2 dpf: Control- 58.25 (SD= 2.6), shp2 MO- 35.75 (SD= 3.2), shp2 MO +LS mRNA= 66.0 (SD= 5.12). p <0.0001 for comparison of shp2 MO to shp2 MO + LS mRNA. Error bars correspond to standard deviation of the mean. |
| Gene: | |
|---|---|
| Fish: | |
| Knockdown Reagents: | |
| Anatomical Term: | |
| Stage: | Day 4 |
| Fish: | |
|---|---|
| Knockdown Reagents: | |
| Observed In: | |
| Stage: | Day 4 |
Reprinted from Developmental Cell, 18(5), Stewart, R.A., Sanda, T., Widlund, H.R., Zhu, S., Swanson, K.D., Hurley, A.D., Bentires-Alj, M., Fisher, D.E., Kontaridis, M.I., Look, A.T., and Neel, B.G., Phosphatase-Dependent and -Independent Functions of Shp2 in Neural Crest Cells Underlie LEOPARD Syndrome Pathogenesis, 750-762, Copyright (2010) with permission from Elsevier. Full text @ Dev. Cell