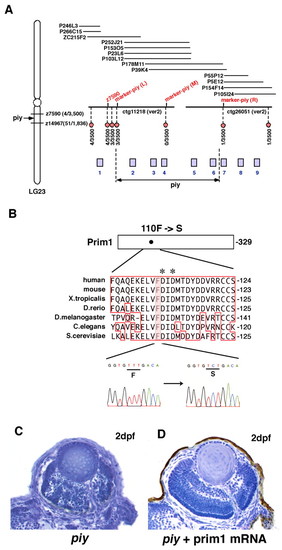
Missense mutation occurs in Prim1 in the piy mutant. (A) Schematic of zebrafish LG23. The piy mutation maps to the genomic region between the SSLP markers z7590 (recombination 4/3500) and z14967 (recombination 51/1836). z7590 maps to the genomic fragment ctg11218, which was shown in the zebrafish genomic database (version 2) released by the Wellcome Trust Sanger Institute. ctg11218 is connected to another genomic fragment, ctg26051, through six overlapping PAC clones. According to the most current version of the zebrafish genomic database, the following nine genes are annotated within this genomic region: 1, zgc:111964 (bHLH transcription factor neuroB); 2, rnd1 (Rho family GTPase 1); 3, prim1 (DNA primase small subunit); 4, zgc:86644 (Tuftelin interacting protein 11); 5, naca (Nascent polypeptide-associated complex alpha); 6, 559664 (EntrezGene) (intracellular membrane-associated calcium-independent phospholipase A2 gamma); 7, 55959 (EntrezGene) (Neurofascin); 8, zgc:65788 (Chitinase); and 9, zgc:123113 (Peptidase). Red circles indicate polymorphic markers that we examined; their recombination rates are shown beneath the red circles. We identified three polymorphic markers, marker-piy (L) (3/3500), marker-piy (M) (0/3500) and marker-piy (R) (1/3500), two of which flanked the piy mutation. Five candidate genes, rnd1, prim1, zgc:86644 (tfip11), naca and 559664 are located in this flanking region. (B) Missense mutation occurred in the prim1 gene in the piy mutant. Amino acid comparison of Prim1 among human (Stadlbauer et al., 1994), mouse (Prussak et al., 1989), Xenopus tropicalis (GenBank accession BC067914), Danio rerio (GenBank accession NM_201448), Drosophila melanogaster (Bakkenist and Cotterill, 1994), Caenorhabditis elegans (GenBank accession NM_06488) and Saccharomyces cerevisiae (Plevani et al., 1987) is shown. Phenylalanine (F) 110 is conserved among these organisms (highlighted). Aspartic acids (D) marked by asterisks indicate the putative active site residues of Prim1 (Augustin et al., 2001). Sequences of the prim1 cDNA in the wild type (left) and piy mutant (right) are shown beneath. The transversion from T to C replaces phenylalanine (F) 110 with serine (S) in the piy mutant. (C,D) piy mutant retinas injected without (C) and with (D) prim1 mRNA.
|

