- Title
-
Redundant and novel functions of scube genes during zebrafish development
- Authors
- Tran, Q.D., Mirkovic, I., Miles, L.B., Berger, J., Wood, A.J., Ruparelia, A.A., Shehni, S.A., Currie, P.D.
- Source
- Full text @ Dev. Biol.
|
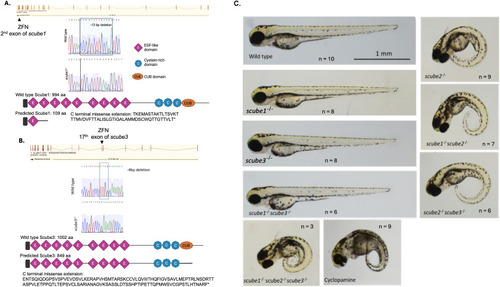
Morphology of the scube mutant family at 3 dpf. (A) Genetic features of scube1 mutants. The scube1 gene comprises 23 exons and encodes a protein of 994 amino acids. ZFN editing targeted the second exon, producing a 13-base pair deletion. This mutation introduces a premature stop codon at position 110. The mutated Scube1 protein is predicted to contain only one EGF domain and no other domains. (B) Genetic features of scube3 mutants. The scube3 gene consists of 22 exons and encodes a protein of 1002 amino acids. ZFN editing targeted exon 17, resulting in a 4-base pair deletion that generates a premature stop codon at position 850. The truncated Scube3 protein is predicted to lack the CUB domain. (C) Morphology of scube mutants at 3 dpf, sample size n = 3–10 embryos. Wild-type larvae at 3 dpf display a straight body axis. Similarly, scube1, scube3, and double mutants (scube1−/−scube3−/−) also maintain a straight body morphology. In contrast, scube2−/−, scube1−/−scube2−/−, scube2−/−scube3−/−, and triple mutants (scube1−/−scube2−/−scube3−/−) exhibit a curved body phenotype, characteristic of Hh signalling disruption. This curved body morphology resembles phenotypes caused by treatment with Cyclopamine, an inhibitor of the Hh pathway. |
|
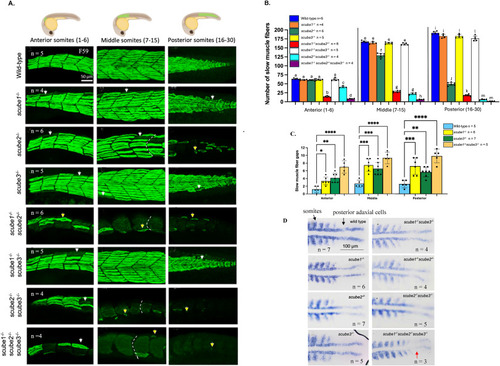
scube mutants display Hh loss-of-function phenotypes. (A) Loss of slow muscle fibres in scube mutant families at 30 hpf. In wild-type zebrafish, slow muscle fibres form a regular array with clearly defined V-shaped somites. In scube1−/−, scube3−/−, and scube1−/−scube3−/−, morphology of slow muscle fibres are comparable to wild-type siblings. In contrast, scube2−/−, scube1−/−scube2−/−, scube2−/−scube3−/−, and the triple scube mutant show severe disruption of slow muscle fibres along the body, including fibre loss (white arrows), abnormal fibre shapes (yellow arrows), and the presence of U-shaped somites (indicated by white dashed lines). The triple mutant shows the most severe phenotype, characterised by the near-complete loss of slow muscle fibres and pronounced abnormalities in the remaining fibres. (B) Slow muscle fibres were quantified in the anterior (somites 1–6), middle (somites 7–15), and posterior (somites 16–30) regions of 30 hpf embryos. In each region, bars labelled with different letters (a–m) represent statistically significant differences. For example, in anterior somites, the number of slow muscle fibres in wild-type, scube1−/−, scube3−/−, and scube1−/−scube3−/− were labelled ‘a' because they are not statistically different from each other; however, they are statistically different from scube1−/−scube2−/− (labelled ‘b'), scube2−/−scube3−/− (labelled ‘c'), and triple scube knockout (labelled ‘d'). Statistical analysis was performed using ANOVA with multiple comparisons and Bonferroni correction. The numbers of slow muscle fibres in scube1−/−, scube3−/−, and double scube1−/−scube3−/− mutants are not different from the wild-type, except in the posterior region of the double mutant, where fibre number is significantly reduced compared to wild-type siblings. Combining scube2 KO with either scube1 or scube3 KO results in a significantly greater reduction in muscle fibres than in single mutants. The triple mutant has the lowest number of slow muscle fibres among all genotypes tested. (C) Fibre gaps in scube1−/−, scube3−/−, and scube1−/−scube3−/− mutants at 30 hpf were counted. Each fibre gap comprised approximately one to four fibres. Statistical analysis showed a significant increase in the number of fibre gaps in each mutant compared to wild-type siblings. Statistical analysis was performed using ANOVA with multiple comparisons and Bonferroni correction. (D) myoD staining in the scube mutant family at 15-somite stage, n = 3 to 7 per mutant. Dorsal views with the head oriented to the left. In wild-type, single, and double scube mutants, myoD expression is expressed in adaxial cells and somites. In the triple mutant (scube1−/−scube2−/−scube3−/−), myoD expression in posterior adaxial cells is lost (red arrow), indicating severe disruption of Hh signalling. |
|
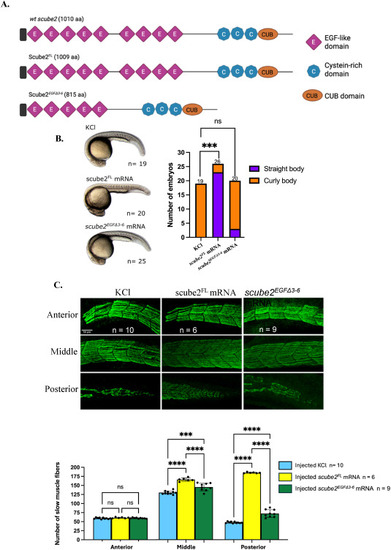
Rescue of scube2−/−phenotype by scube2FLandscube2EGFΔ3-6mRNAs. (A) The Scube2FL protein is predicted to closely resemble the wild-type Scube2 with 9 EGF-like domains, while the Scube2EGFΔ3-6 peptide only has 5 EGF-like domains. (B) Morphological analysis of scube2−/− larvae at 30 hpf following 0.2 M KCl injection showed that all scube2−/− larvae exhibited a curled body axis (19/19). Injection of scube2FL mRNA restored a straight body axis in most larvae (23/26), while injection of scube2EGFΔ3-6 mRNA did not rescue normal morphology, with most larvae remaining curled (17/20). The difference between the control group (injected with 0.2 M KCl) and the scube2FL mRNA group was statistically significant (Fisher's exact test with Bonferroni correction, p < 0.001). No significant difference was observed between the control and scube2EGFΔ3-6 mRNA groups (p = 0.681). (C) Slow muscle immunostaining in scube2−/− embryos at 30 hpf following rescue with scube2FL or scube2EGFΔ3-6 mRNAs revealed that injection of either scube2FL or scube2EGFΔ3-6 mRNA led to a significant increase in slow muscle fibre number compared to the control. The increase was greater in scube2−/−embryos rescued with scube2FL than with scube2EGFΔ3-6 (ANOVA with Bonferroni correction, p < 0.0001). |
|
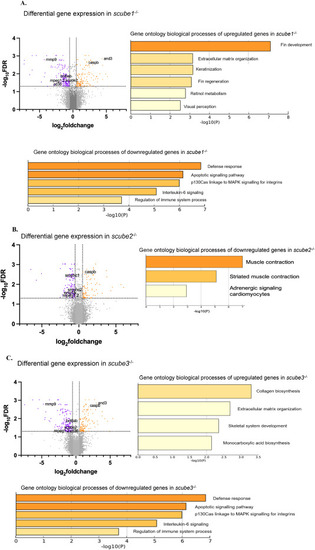
RNA seq analysis of scube1−/−, scube2−/−and scube3−/−embryos at 30 hpf. (A) RNA sequencing of scube1 mutants revealed upregulation of processes related to fin development and regeneration, extracellular matrix formation, keratinisation, and eye development. Conversely, there is a downregulation of processes associated with the immune response system (response to bacterium, interleukin-6 signalling, response to wounding, platelet degranulation Herpes infection), apoptotic signalling and response to estrogen. (B) RNA sequencing of scube2−/− zebrafish mutants indicated downregulation of processes linked to muscle contraction and adrenergic signalling in cardiomyocytes. (C) RNA sequencing of scube3−/− zebrafish mutants revealed upregulation of processes related to collagen synthesis, extracellular matrix formation, and skeletal system development. Conversely, downregulated processes include those involved in the defence response system, specifically interleukin-6 signalling and the regulation of immune system processes. Yellow bars indicate lower significance or enrichment, while orange bars indicate higher values. FDR cutoff = 0.05. Volcano plots displaying differential gene expression between scube mutants and wild-type zebrafish. The x-axis shows log2 fold change in gene expression, and the y-axis indicates - log10 FDR. Each point represents an individual gene. Genes with absolute log2 fold change greater than 0.5 and FDR less than 0.05 are highlighted: upregulated genes are orange, downregulated genes are purple, and non-significant genes are grey. |
|
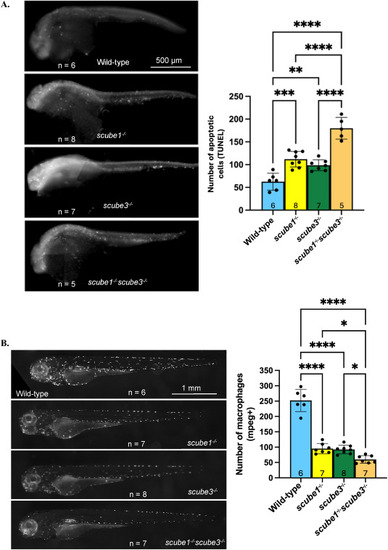
Increased apoptosis and reduced macrophage numbers in scube mutants. (A) The TUNEL assay revealed a greater number of apoptotic cells in scube1−/−, scube3−/−, and scube1−/−scube3−/− compared with wild-type at 30 hpf. Double mutants for scube1 and scube3 showed significantly higher numbers of apoptotic cells than either single mutant. ANOVA F(3,24) = 107.9, p < 0.0001. (B) Macrophage numbers decrease in scube mutants. Tg(mpeg1:Gal4-FF)gl25/Tg(UAS-E1b:nfsB.mCherry)c264 reporter line was used to label macrophages. Live imaging of zebrafish at 3 dpf showed fewer macrophages in scube1−/−, scube3−/−, and the double mutant scube1−/−scube3−/− compared to controls. The double mutant displayed a further reduction in macrophage numbers compared to either single mutant. ANOVA, F (3, 22) = 41.26, p < 0.0001. |
Reprinted from Developmental Biology, , Tran, Q.D., Mirkovic, I., Miles, L.B., Berger, J., Wood, A.J., Ruparelia, A.A., Shehni, S.A., Currie, P.D., Redundant and novel functions of scube genes during zebrafish development, , Copyright (2025) with permission from Elsevier. Full text @ Dev. Biol.