- Title
-
Impaired Detoxification of Trans, Trans-2,4-Decadienal, an Oxidation Product from Omega-6 Fatty Acids, Alters Insulin Signaling, Gluconeogenesis and Promotes Microvascular Disease
- Authors
- Qian, X., Klatt, S., Bennewitz, K., Wohlfart, D.P., Lou, B., Meng, Y., Buettner, M., Poschet, G., Morgenstern, J., Fleming, T., Sticht, C., Hausser, I., Fleming, I., Szendroedi, J., Nawroth, P.P., Kroll, J.
- Source
- Full text @ Adv Sci (Weinh)
|
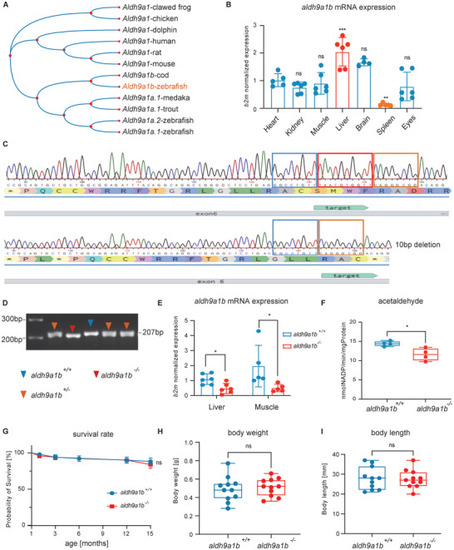
Generation and validation of |
|
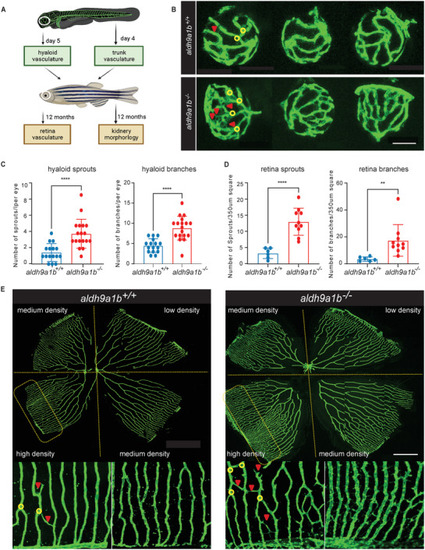
Alteration of the retinal vasculature in |
|
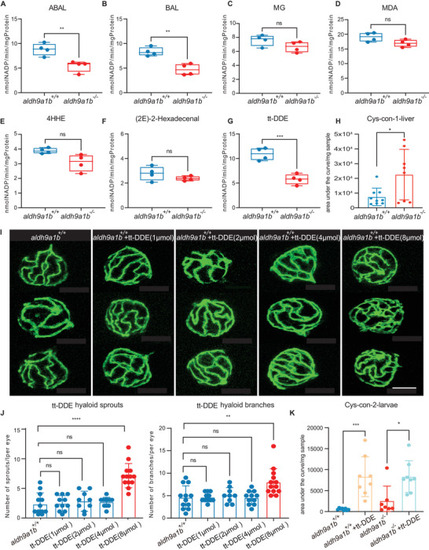
The link between tt‐DDE, Aldh9a1b, and angiogenesis (A‐G) Aldh enzyme activity was significantly decreased using substrate ABAL (A), BAL (B), and tt‐DDE (G), but unaltered with substrate MG (C), MDA (D), 4HHE (E) and 2(E)−2‐hexadecenal (F) in |
|
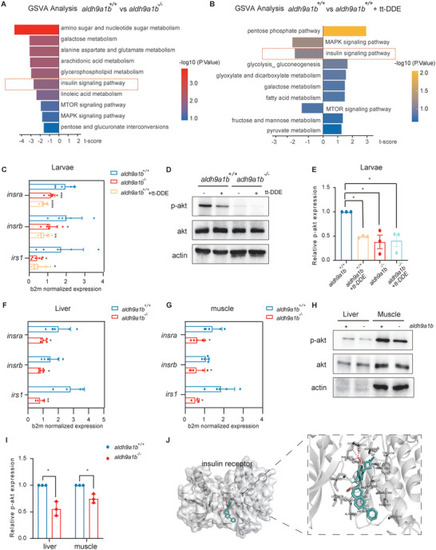
Downregulated insulin receptor signaling pathway in |
|
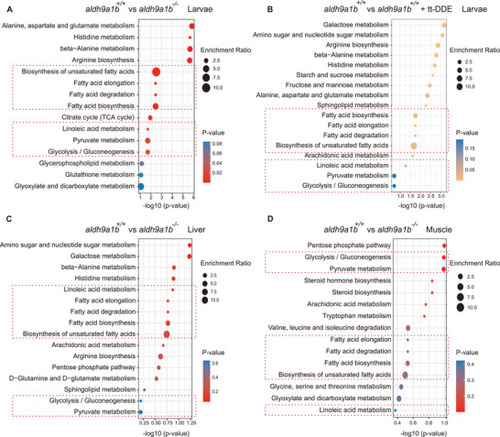
Altered glucose metabolism in |
|
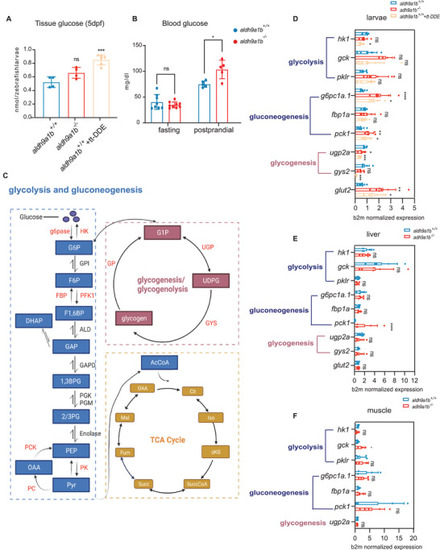
Impaired glucose homeostasis in |
|
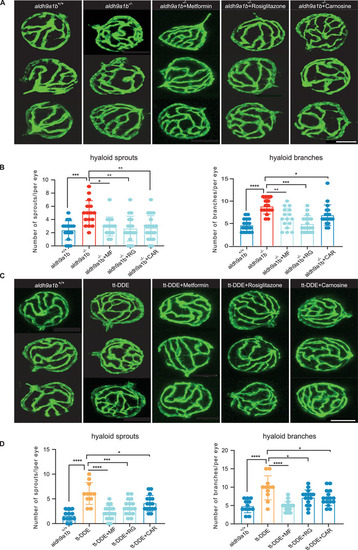
Altered angiogenic hyaloid vasculature caused by |