- Title
-
Determining zebrafish dorsal organizer size by a negative feedback loop between canonical/non-canonical Wnts and Tlr4/NFκB
- Authors
- Zou, J., Anai, S., Ota, S., Ishitani, S., Oginuma, M., Ishitani, T.
- Source
- Full text @ Nat. Commun.
|
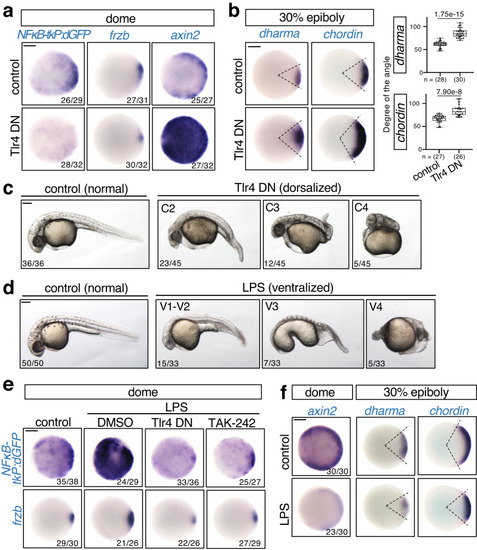
Rel/NFκB negatively regulates dorsal organizer formation. |
|
Specific |
|
Rel negatively regulates Wnt/β-catenin signaling. Wnt/β-catenin signaling is enhanced by |
|
Rel directly activates a secreted Wnt antagonist Frzb to restrict Wnt/β-catenin-mediated dorsal organizer formation. |
|
Tlr4 activates NFκB to stimulate |
|
β-catenin stimulates Wnt5b-mediated Tlr4/NFκB activation. |
|
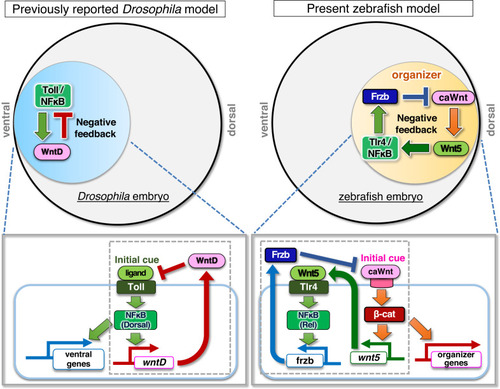
Negative feedback loop between canonical/non-canonical Wnts and Tlr4/NFκB determines the precise size of zebrafish dorsal organizer. Model of the role of Tlr/NFκB signaling in the initiation of embryonic DV axis formation. In |