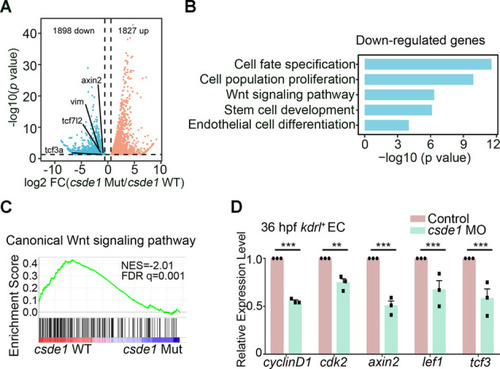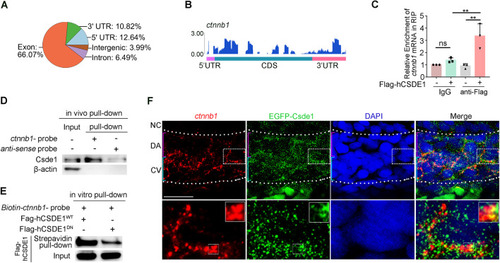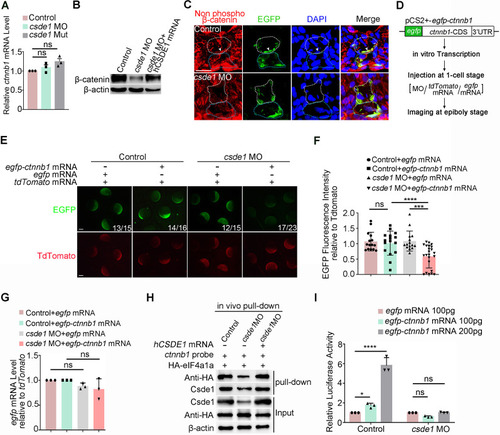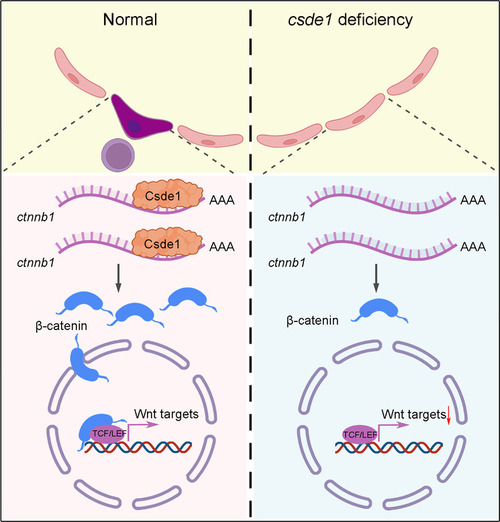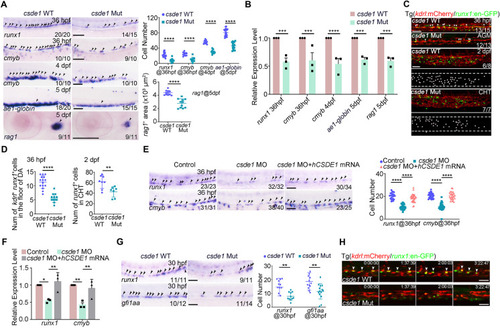
HSPC generation is impaired in csde1 mutants. (A) Expression of HSPC markers runx1 and cmyb (arrowheads) in the AGM region at 36 hpf, cmyb (arrowheads) in the CHT region at 4 dpf, erythroid marker ae1-globin and lymphoid marker rag1 (arrowheads) in the thymus region at 5 dpf in csde1 mutants and WT by WISH, with quantification (right panels). (B) qPCR analysis of runx1, cmyb, ae1-globin and rag1 in csde1 mutants and WT at 36 hpf, 4 dpf or 5 dpf. (C,D) Confocal imaging showing kdrl+runx1+ HECs (white arrowheads) in the AGM region at 36 hpf and runx1+ HSPCs in the CHT region at 2 dpf in csde1 mutants and WT (C) and quantification (D). Dashed lines in C outline the CHT region. (E,F) WISH (E, with quantification, right panel) and qPCR (F) analysis showing that the expression of runx1 and cmyb (arrowheads) at 36 hpf was rescued by hCSDE1 mRNA, compared with csde1 morphants. (G) Examination of the HEC marker runx1 and gfi1aa expression in WT and csde1 mutants at 30 hpf by WISH, with quantification (right panel). (H) Snapshot of EHT (arrowheads) in csde1 mutants and siblings. Data are mean±s.d. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (two-tailed unpaired Student's t-test). n≥3 replicates. Numbers indicate the number of embryos with respective phenotype/total number of embryos analyzed in each experiment (A,C,E,G). Scale bars: 100 μm (A,E,G); 50 μm (C,H).
|