- Title
-
Functional analysis of germline VANGL2 variants using rescue assays of vangl2 knockout zebrafish
- Authors
- Derrick, C.J., Szenker-Ravi, E., Santos-Ledo, A., Alqahtani, A., Yusof, A., Eley, L., Coleman, A.H.L., Tohari, S., Ng, A.Y., Venkatesh, B., Alharby, E., Mansard, L., Bonnet-Dupeyron, M.N., Roux, A.F., Vaché, C., Roume, J., Bouvagnet, P., Almontashiri, N.A.M., Henderson, D.J., Reversade, B., Chaudhry, B.
- Source
- Full text @ Hum. Mol. Genet.
|
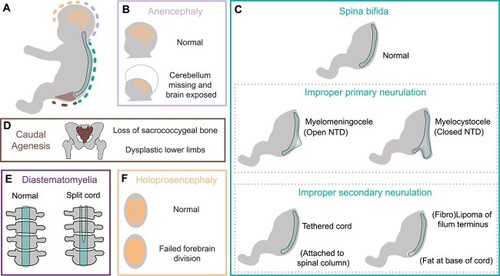
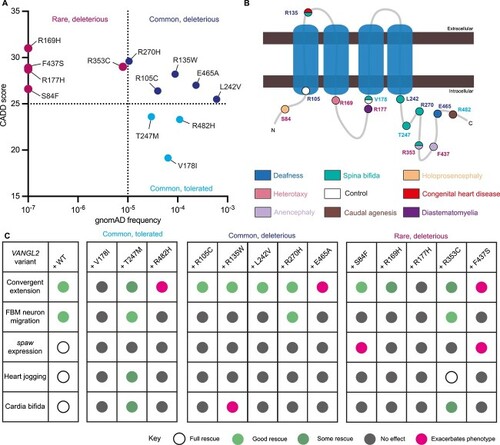
Congenital malformations associated with |
|
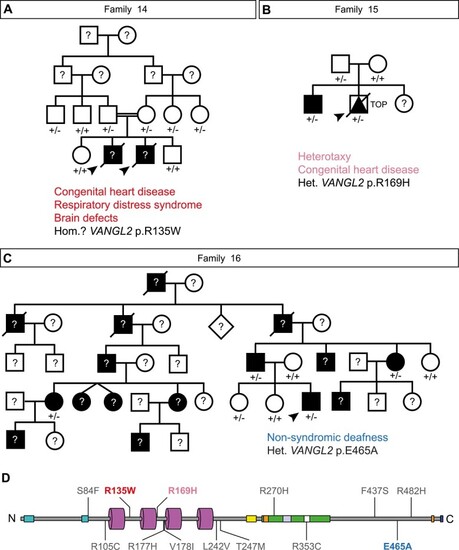
Collation of clinical data relating to historical |
|
Identification of new |
|
PHENOTYPE:
|
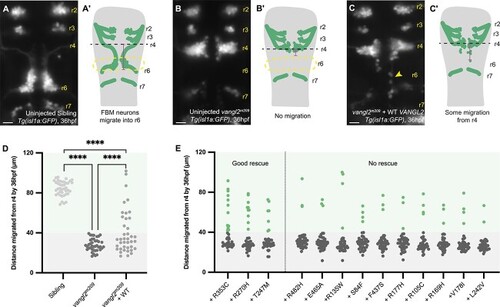
|
Motor neuron migration cannot be rescued by most |
|
Asymmetric EXPRESSION / LABELING:
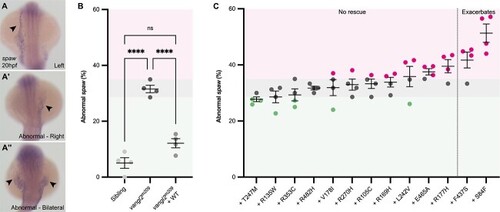
PHENOTYPE:
|
|
Heart tube formation and positioning is sensitive to loss of |
|
Summary of |