- Title
-
Syntaxin 18 Defects in Human and Zebrafish Unravel Key Roles in Early Cartilage and Bone Development
- Authors
- Guillemyn, B., De Saffel, H., Bek, J.W., Tapaneeyaphan, P., De Clercq, A., Jarayseh, T., Debaenst, S., Willaert, A., De Rycke, R., Byers, P.H., Rosseel, T., Coucke, P., Blaumeiser, B., Syx, D., Malfait, F., Symoens, S.
- Source
- Full text @ J. Bone Miner. Res.
|
Pedigree of the family, radiographs of the proband, structure of the human STX18 gene and protein, and overexpression of human wild‐type (WT) and p.(Arg10Pro) STX18 proteins. (A) Pedigree of the Turkish family. The proband (II:1) is indicated with an arrow, and asterisks denote family members available for molecular testing. The parents are obligate heterozygous carriers of the identified homozygous STX18 variant in the proband (c.29G > C). (B, C) Radiographs of II:1 at 26 weeks of gestation reveal multiple fractures, thin ribs, abnormal cartilage in the spine (arrowhead), short and cracked femur (arrowhead), tibial bowing (arrowhead), irregular mandible (arrowhead), and severe cranial undermineralization (arrowhead). (D) Human STX18 consists of 10 exons and comprises 123.08 kb on chromosome 4; human STX18 protein (Q9P2W9) domain structure includes cytoplasmic, transmembrane (TM), and vesicular (ves) domains. The identified homozygous p.(Arg10Pro) variant is part of the N‐terminal α‐helical cytoplasmic domain. Numbers indicate amino acid residues. (E) Overexpression of WT and mutant STX18 proteins lead to stable protein products in HeLa and Saos‐2 cells. |
|
Generation and characterization of mosaic stx18 zebrafish. (A) Zebrafish stx18 comprises 11 exons, is 5.16 kb long, and maps to chromosome 4. The stx18 protein (Q4VBI7) has similar domains as its human counterpart. The location of the CRISPR/Cas9 target sites are indicated and are located in exons 2 (design 2–validation purposes), 3 (design 3–validation purposes), and 4 (design 1–main article). (B) The majority of generated indels (98% of alleles) in the mosaic uppercut18 zebrafish model (“uppercut18 crispants”) result in a premature termination codon (PTC, 73% out‐of‐frame). (C) Uppercut18 zebrafish have a curved notochord (arrowhead), display cranial distortions (with severely deformed lower jaw, arrowhead), and have malformed pectoral fins (arrowheads) and an underdeveloped swim bladder that is not inflated (highlighted with a dotted white line and arrowhead). (D) Standard length of scrambled (n = 96) and uppercut18 (n = 86) zebrafish. The mean of data points is indicated, and an unpaired t test was used to determine significance. Each data point represents the measurement from 1 zebrafish, with highlighted median value (line). Two‐tailed unpaired t test; ****p < 0.0001. (E) Head length of scrambled (n = 64) and uppercut18 (n = 64) zebrafish. The mean of data points is indicated, and an unpaired t test was used to determine significance. Each data point represents the measurement from 1 zebrafish, with highlighted median value (line). Two‐tailed unpaired t test; ****p < 0.0001. (F) Survival curve of scrambled (n = 125) and uppercut18 (n = 127) zebrafish. All uppercut18 zebrafish died by 11 dpf. (G) Lateral (top) and ventral (bottom) views of Alcian blue–stained uppercut18 zebrafish reveal malformed Meckel's cartilage (mk), a smaller ethmoid plate (em), malformed ceratohyals (ch) and ceratobranchial pairs 1 to 5 (cb), and malformed pectoral fins (arrows). (H) Ventral view of Alizarin red–stained cranial skeleton reveals a generalized delay in ossification of the intramembranous and endochondral skeleton (p = parasphenoid; n = notochord; c = cleithrum; cb = ceratobranchial 5; o = opercle; hm = hyomandibular; br3 = branchiostegal ray 3) with absence of the entopterygoid (en). Images in C, G, and H are oriented to the left; scale bar = 1 mm. For C, G, and H, 20 5‐dpf scrambled and crispant zebrafish were analyzed and representative samples are shown. |
|
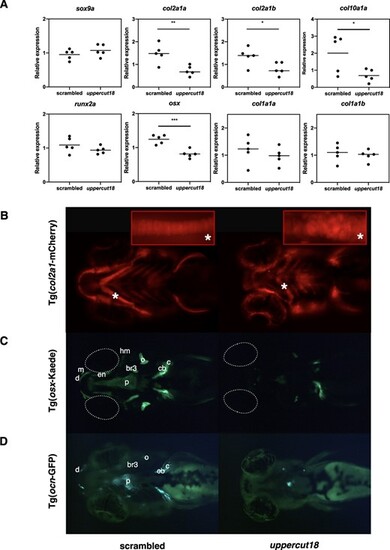
Five‐dpf‐old uppercut18 zebrafish have impaired bone development. (A) mRNA levels of sox9a, runx2a, col1a1a, and col1a1b are unaltered in uppercut18 zebrafish, but transcriptional levels of col2a1a, col2a1b, osx, and col10a1a are downregulated in uppercut18 zebrafish. Each data point represents the mean of 2 technical measurements from 5 biological replicates, with highlighted mean value (line). Two‐tailed unpaired t test; *p < 0.05, **p < 0.01, and ***p < 0.001. (B) Defective cartilage development with associated impaired chondrocyte stacking (asterisk) is noted in uppercut18 zebrafish when using Tg(col2a1:mCherry) reporters. (C, D) Developmental delay of uppercut18 zebrafish in osterix (Tg[osx:Kaede]) and osteocalcin (Tg[ocn:GFP]) reporters. (C) Osterix is less expressed at the level of the ceratobranchial 5 (cb), cleithrum (c), opercle (o), and parasphenoid (p) structures; signal at the branchiostegal ray 3 (br3), hyomandibular (hm), entopterygoid (en), maxilla (m), and dentary (d) cranial head bone structures is absent in uppercut18 zebrafish. Dotted white lines represent the location of the eyes. (D) Osteocalcin is only expressed on a small part of the parasphenoid (p), whereas signal at the ceratobranchial 5 (cb), cleithrum (c), opercle (o), branchiostegal ray 3 (br3), and dentary (d) cranial head bone structures is absent in uppercut18 zebrafish. For B–D, 20 5‐dpf scrambled and crispant zebrafish were analyzed and representative samples are shown. |
|
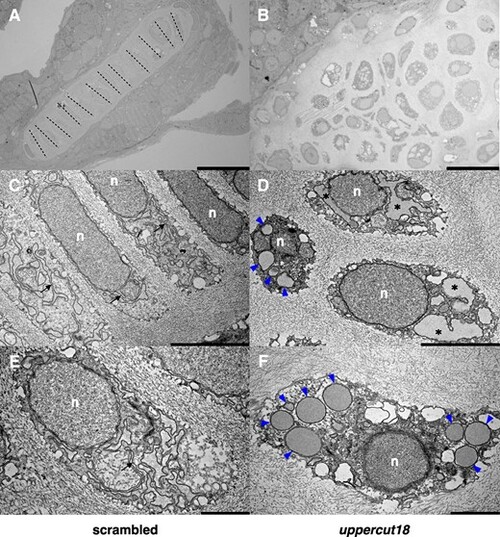
Five‐dpf‐old uppercut18 zebrafish have compromised chondrocyte morphology and function. Electron micrographs of scrambled zebrafish (A, C, E) show cuboidal (polarized) chondrocytes that are in well‐organized stacks (dashed lines) and contain dense ER membranes that are well developed and organized (arrows), whereas uppercut18 zebrafish (B, D, F) are associated with irregularly formed and organized chondrocytes, distended ER (asterisks), and large amounts of electron‐dense material accumulating in the rough ER (blue arrowheads). n = nucleus. Scale bars (A–F) = 20, 5, and 2 μm, respectively. For A–F, 10 5‐dpf scrambled and crispant zebrafish were analyzed and representative samples are shown. |
|
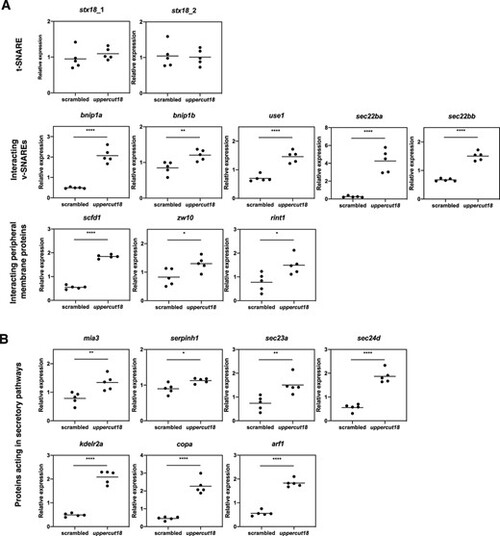
Five‐dpf‐old uppercut18 zebrafish are associated with altered transcriptional levels of SNARE complex members, protein interacting components, and components of the secretory pathway. (A) Transcriptional levels of stx18 are unaltered in uppercut18, but SNARE protein members including bnip1a, bnip1b, use1, sec22ba, and sec22bb are strongly upregulated, as well as the peripheral membrane protein components scfd1, zw10, and rint1. (B) Upregulated mRNA levels in uppercut18 zebrafish of all components investigated involved in early, mid, late anterograde, and retrograde secretory pathways (mia3, serpinh1, sec23a, sec24d, kdelr2a, copa, and arf1). Each data point represents the mean of 2 technical measurements from 5 biological replicates, with highlighted mean value (line). Two‐tailed unpaired t test; *p < 0.05, **p < 0.01, and ****p < 0.0001. |